 |
| Books and articles on the evolution of the immune system. Click to enlarge. |
by Nick Matzke
This webpage contains the same articles and books used during the Behe cross-examination that are listed in the Supplementary Material for Bottaro et al. (2006). Here, the citations have been placed in chronological order, and each reference is annotated with a blue box containing a short summary of the significance of the paper, and if relevant, significant quotes from the paper.
Introduction
The purpose of this annotated bibliography is to give the nonspecialist reader some sense of the scholarly weight of the evolutionary immunology literature (here is another method). In particular, this bibliography focuses attention on the scientific work that developed, tested, and established the "transposon hypothesis" for the origin of receptor rearrangement in the adaptive immune system. Contrary to the impression one might get from the ID literature, the transposon hypothesis was not idle speculation, it was not thought up yesterday over breakfast, it is not particularly vague, it is not untestable, and it is not scientifically useless. In contrast, by reading through the bibliography in chronological order, we can see that the transposon hypothesis was explicitly proposed and published in top journals, it was carefully and seriously discussed for decades in the professional literature, it has inspired a very productive research program (both experimental and comparative), it has been tested with diverse evidence by researchers working in many different labs, and it has been dramatically confirmed.
In short, it is a classic case of serious, advancing evolutionary science. Without ever getting mentioned in a newspaper article or cable news show, hundreds of PhD scientists have devoted their careers to working out how and why the immune system evolved. Any one of these researchers has quietly produced more research results than the entirety of the intelligent design movement with their collection of op-eds, webpages, in-house publications, books published by InterVarsity Press, one or two books by slightly more rigorous publishers but with dubious peer-review, and one or two review articles slipped into obscure journals. Unlike "intelligent design" proponents, evolutionary immunologists repeatedly published their key work in top journals. For some reason, they have not felt the need to write law review articles, lobby school boards and legislatures to get their view taught, or muck around with state science standards, because they know that scientific hypotheses succeed by convincing other scientists through research.
For more along these lines, see Bottaro et al. (2006) and the
Why the Immune System?
The decision to make the evolution of the immune system a key example in both Kenneth Miller's direct testimony and Michael Behe's cross-examination was based on several factors. First, among the major systems that Behe discusses in Darwin's Black Box, such as the eukaryotic cilium, the bacterial flagellum, the blood-clotting cascade, and the immune system, Behe makes particularly florid claims about the absence of literature on the origin of the immune system -- for example:As scientists we yearn to understand how this magnificent mechanism came to be, but the complexity of the system dooms all Darwinian explanations to frustration. (Darwin's Black Box, p. 139)
We can look high or we can look low, in books or in journals, but the result is the same. The scientific literature has no answers to the question of the origin of the immune system. (Darwin's Black Box, p. 138)These are claims crying out for refutation. As it happens, there is probably more scientific literature directly on the evolution of the immune system than on the other three systems put together (we can chalk this up to (1) the long tradition of evolutionary and comparative studies in immunology; (2) the age of the discipline (going back to the 1800's); and (3) the massive amount of medical research money available for studies of the immune system, for obvious reasons). So, while Behe makes various mistakes on the other systems, many also discussed at trial, he is most dramatically wrong about the immune system literature.
Second, although the immune system is very complex, everyone is familiar with (a) its importance, (b) vaccinations and immunity to disease, and (c) many people have heard of antibodies. So it is actually not so difficult for a nonspecialist, such as a lawyer or judge, to realize the importance of immune system research.
Third, those in the community of "creationism watchers" knew that the ID advocates were particularly vulnerable on the immune system, given their past flailings in response to internet challenges. Some of this is mentioned in Bottaro et al. (referencing these Panda's Thumb posts: [1] -- [2]) and Michael Behe's weak response. For an earlier episode often recalled by creationism watchers see this celebrated discussion on an ID bulletin board, where several pro-evolution posters challenged several leaders of the ID movement to admit that Behe's 1996 claim was wrong.
Fourth, Behe's dismissal of several famous articles on evolutionary immunology during his deposition also increased confidence that he was not at all familiar with the literature, and instead would brush off any challenges as not meeting his requirement for infinite detail.
Fifth, plaintiffs' attorney Eric Rothschild had the strength of will to continue with the immune system gambit, despite Nick Matzke's early attempt at explaining the evolution of V(D)J recombination to him on a whiteboard:
 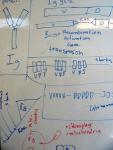 |
| The beginnings of the immune system "battle plan" for the Behe cross-examination. Whiteboard drawings from May 2005, NCSE office. Click to see larger images. |
Scientific Background
The annotated bibliography will not make much sense without a little background understanding of immunology. If you can understand this basic terminology, you should be well on your way to understanding the transposon hypothesis:
- Adaptive immune system (AIS): The portion of the immune system that generates receptors that specifically recognize and bind to a particular pathogen. This system has "memory", so that if a pathogen returns, it will be recognized much more quickly the second time around. Vaccines expose a person to a killed or weakened form of a pathogen, or key pieces of a pathogen. This "trains" the immune system to recognize common pathogens such as smallpox or polio, so that immunity can be developed without the person having to catch and fight off the actual disease. The adaptive immune response is restricted to jawed vertebrates (i.e., all vertebrates except lampreys and hagfish), and therefore evolved 450-500 million years ago in the common ancestor of cartilagenous fish (sharks and rays) and other "fish" (bony fish, including tetrapods). (Note: there is increasing evidence of different forms of adaptive responses in other organisms.)
- Innate immune system: The portion of the immune system that uses general, nonspecific responses to fight pathogens. For example, a receptor molecule called TLR5 recognizes flagellin, the major component of bacteria flagella and a common conserved feature. The innate immune system is the first line of defense, and many of its features are shared between vertebrates and invertebrates
- Immunoglobulins (Igs), also called antibodies: These are the receptor proteins of the immune system. They have a "Y" shape, where the top of the "Y" recognizes foreign molecules called antigens. The genes that encode immunoglobulins are composed of four basic kinds of segments: V (variable), D (diversity), J (joining), and C (constant). Vertebrates have hundreds of different copies of V, D, and J gene segments. Through a process called V(D)J recombination, different copies of these segments are spliced together, producing billions of different unique immunoglobulin receptors. Most of these receptors will not recognize a particular pathogen, but a few of them will, even if it is a completely novel pathogen.
- Recombination Activating Gene (RAG): A RAG is a gene that codes for a RAG protein (the names of genes are italicized, and the corresponding proteins are not; sometimes PDF and HTML documents will be missing this formatting however). RAG proteins have the ability to recognize specific DNA sequences at two locations called Recombination Signal Sequences (RSSs), bring the pieces together, and cut the DNA at the RSS sites. DNA repair enzymes then repair the DNA and join the two segments, originally distant from each other, together. In the vertebrate immune system, two RAGs, RAG-1 and RAG-2, cooperate in this process.
- Transposon: A transposon, or transposable genetic element, is a "jumping gene." The transposon is usually a gene that codes for a protein. This protein, in turn, recognizes certain signal sequences (sound familiar?) in the transposon DNA allowing it to snip out the transposon DNA and transplant it someplace else. There are many different types of transposons with various degrees of relationship to each other, and they are found in all groups of organisms. Mutations can "break" transposons, resulting in "fossil" transposons that are thought to usually be "junk DNA" (although it may play some non-coding, structural role in cells). About 3% of the human genome is composed of the remains of DNA transposons, and about half of the genome is made of various other kinds of copied repetitive DNA.
With that terminology in hand, we can discuss the transposon hypothesis. The transposon hypothesis is based on similarities between transposons and RAGs, and suggests that RAG-1 and RAG-2, and the RSSs, are actually descended from "free-living" transposons. The model states that in an early vertebrate, already equipped with an innate immune system, a transposon inserted into a non-rearranging receptor gene. When this receptor gene was expressed, the transposon was activated, and snipped itself out of the receptor. However, because this excision process was inexact, the resulting receptor would be variable, and therefore some copies of the receptor protein would be better able to recognize new or mutated pathogens. Natural selection would therefore spread this variant. An extended process of gene duplication and diversification would elaborate this basic system into the modern V(D)J recombination system.
The transposon hypothesis suggests a number of specific observations that would corroborate the model if found:
- sequence similarities between V(D)J RSSs and transposon recognition sequences
- RAGs should operate by mechanisms similar to transposase mechanisms
- RAGs might still be able to perform the same functions that transposons perform, such as DNA excision and insertion
- immunoglobulins should have relatives that are non-rearranging receptors
- if RAGs are descended from transposons, then "free-living" transposon relatives of RAG might still exist "in the wild"
All of these observations have been published in the last 10 years. The crowning achievements, and the easiest successes to understand, were the identification of transposon relatives of RAG-1 in sea urchins, lancelets, and cnidarians (Kapitonov and Jurka, 2005) and a RAG1-RAG2 homolog serving a non-immune function in sea urchins (Fugmann et al. 2006).
If the above, very short, summary did not completely sink in, other resources are available. For an excellent moderate-length introduction to evolutionary immunology, please see Matt Inlay's online article, "Evolving Immunity." Pay particular attention to the graphics. Next, read our Nature Immunology essay for a brief update on the scientific progress of the transposon hypothesis.
See also these Panda's Thumb blogposts by Matt Inlay or Andrea Bottaro: "New discovery of missing link between adaptive immune system and transposons," "The Revenge of Calvin and Hobbes," and "Behe's meaningless complexity." Once you have given those sources a try, take several minutes to work through Figure 1 and Figure 2 of Lewis and Wu's (2000) commentary article, "The Old and the Restless", published in the Journal of Experimental Medicine. Figure 1 is a model of how receptor rearrangement works in modern organisms. Figure 2 is a model of how it evolved.Another introductory survey can be found in the immunology textbook, Immunobiology: the immune system in health and disease. The 5th edition, Janeway et al. (2001), is freely available online in PubMed's textbooks collection. Chapter 4 is "The Generation of Lymphocyte Antigen Receptors" (sections 1, 2, 3, 4, 5). The Afterword to the book, "Evolution of the Immune System: Past, Present, and Future", is written by Janeway, and summarizes the state of knowledge as of 2001 on the evolution of the innate immune system and evolution of the adaptive immune response. The transposon hypothesis naturally plays a large role in the latter.
Hopefully, after you have worked through this material you can re-read the above summary and make some headway.
How the Bibliography was Assembled
This bibliography was originally developed for the Behe cross-examination in the Kitzmiller case, discussed in Bottaro et al. (2006). The annotations are based on notes that were taken while the bibliography was being assembled (in an all-night session the weekend before Behe testified, it must be said). The annotations have subsequently been revised and updated.
While assembling the exhibit, it was very important to make sure it was not vulnerable to some of the critiques that can be leveled against lists of articles that are uncomprehendingly culled from automated literature searches. When one searches online databases for publications that involve the keywords "immune system" and "evolution" -- two immense research topics -- one will get results on a wide variety of topics. These include:
- the evolution of disease organisms
- the evolution of modern populations in response to diseases
- the evolution of immune system cells when an organism's adaptive immune response is triggered (such as when someone is vaccinated)
- the use of customized antibodies for innumerable biomedical purposes
- the "evolution" of chemical reactions or biochemical reactions (this is a nonbiological definition of evolution, for example, the "evolution" of hydrogen gas during hydrolysis)
- clinical research on AIDS, allergies, etc.
- the evolution of the immune system within closely-related organisms -- e.g., mammals, birds, or tetrapods
- "deep" comparative immunology between vertebrates and invertebrates
- the selective forces driving immune system evolution and receptor diversity
- the origin of the innate immune system
- the origin of the adaptive immune system
Only the last four topics directly address the evolutionary origin of the immune system, although many of the other topics can have some relevance. Among these four topics, this bibliography focused mostly on topic #11, and included a smattering of work on topics #8, 9, and 10. Within topic #11, the origin of the V(D)J recombination system was emphasized, as it is widely seen as the "key" system, the most remarkable feature of adaptive immunity, and the biggest evolutionary "puzzle" according to ID advocates.
This bibliography also focused on the review literature rather than the research literature. First, the research literature is mostly opaque to nonspecialists, and the implications of findings are made much clearer in the review literature. Second, at his deposition, Michael Behe expressed the opinion that if scientists had made any progress worth talking about in evolutionary immunology, he would have expected to see it in the review literature. A few of the most famous research articles were included, but most of the articles are articles reviewing the research literature -- many of the review articles cite several hundred other articles.
This literature collection should not be considered comprehensive or complete -- it is merely a sample of the literature on the evolution of the adaptive immune system, focusing on V(D)J recombination, with a small sampling of literature on the evolution of innate immunity, MHC, large-scale comparative immunology, etc. For some idea of how much more scientific literature there is on this topic, see the longer unannotated bibliography.
The Larger Context
Matt Inlay (author of "Evolving Immunity" and coauthor of Bottaro et al. 2006) points out that Michael Behe's book Darwin's Black Box was ironically timed. In 1996, the year Darwin's Black Box was was published, research on V(D)J recombination was in the midst of a dramatic upswing:

|
| Research progress on V(D)J recombination over the last 25 years. Results of a PubMed search on the term "V(D)J recombination", plotted as a function of time (blue dots). Numbers of articles per year are shown on the y-axis, and the years of publication on the x-axis. Publications for 2006 were projected based on Jan-Apr numbers. Major breakthroughs in the transposon model are listed in text boxes. The vertical line denotes 1996, the year Darwin's Black Box was published. The trendline was calculated via moving average with a period of 3. |
| Graph and caption by Matt Inlay. |
Inlay has added to the graph boxes showing the chronology of the major research findings supporting the transposon model. The viewer can see that they came fast and furious after 1996, almost as if on cue.
Major Themes
The story told by this bibliography, and by Ken Miller's testimony in Kitzmiller, and by Bottaro et al. (2006), is basically of the proposal, development, testing, and confirmation of Sakano et al.'s 1979 suggestion in Nature that V(D)J recombination might have emerged when a transposon -- a piece of DNA that codes for a protein that can chop out that piece of DNA and insert it somewhere else -- inserted into the middle of a receptor gene. This hypothesis has been gathering steam for awhile, and really took off in the last ten years.
A subplot in the story is how evolution has played a key role in immunology throughout its development. Sakano et al.'s hypothesis was not born in a vacuum; it occurred in a field where comparative immunology, informed by evolutionary modeling, had been a core part of the discipline for a hundred years, going back to the work of Metchnikoff in the late 1800s.
If one reads chronologically through the bibliography, one will see the researchers themselves telling the same story, and citing the same papers that have been repeatedly cited in the various Behe immune system rebuttals. But you will also see some rather remarkable comments about Sakano et al.'s hypothesis. Some of the best ones will be highlighted.
For example, as the transposon hypothesis began to pick up steam in the mid-1990's, it did attract some critisms. E.g., Lewis and Wu (1997) wrote,It is nonetheless worth noting that in spite of various similarities, there are fundamental differences between transposition and V(D)J recombination. For one, there are no reports of a transposase (mutant or otherwise) that is able to mediate site-specific inversion. Conversely, it has never been demonstrated that V(D)J recombination can cause the integration of one piece of DNA into another. (Lewis and Wu 1997, p. 162)However, the very next year, two labs discovered that the RAG proteins actually could cause the integration reaction. In a research article in Nature entitled "Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system", Agrawal et al. write in their abstract,
Here we show that RAG1 and RAG2 together form a transposase capable of excising a piece of DNA containing recombination signals from a donor site and inserting it into a target DNA molecule. [...] The results support the theory that RAG1 and RAG2 were once components of a transposable element, and that the split nature of immunoglobulin and T-cell-receptor genes derives from germline insertion of this element into an ancestral receptor gene soon after the evolutionary divergence of jawed and jawless vertebrates.In a commentary article in the same issue of Nature, Plasterk (1998) said, "the authors show that the RAG1 and RAG2 proteins (which mediate V(D)J joining) can still catalyse a full transposition reaction. A similar result has been independently obtained by Martin Gellert and co-workers, and is reported in tomorrow's issue of Cell." This second article was Hiom et al. (1998). In 1999, Schatz reviewed the progress of Sakano et al.'s transposon hypothesis and pronounced it a "prophetic hypothesis":
When examined in their germline configuration, the RSSs flanking V and J constitute an inverted repeat, much like that found at the end of transposons, and this realization led to the prophetic hypothesis, in 1979, that insertion of a transposable element into an ancient receptor gene exon was responsible for the generation of split antigen receptor genes during evolution ([Sakano et al., 1979]). (Schatz 1999, p. 170, bold added)Also in 1999, Susanna Lewis -- the same Lewis as Lewis and Wu (1997) -- engaged in a little prophesy of her own. If the RAGs evolved from transposons, she said, it would be really handy to find one:
It would be extremely useful if a contemporary version of the original RAG transposon could be identified. A distant cousin, with credentials, would greatly facilitate any attempts to reconstruct the lost history between the time the first RAG element took up residence in the vertebrate genome and the emergence of a developmental recombination system. (Lewis 1999, p. 65)Six years later, exactly this was reported by Kapitonov and Jurka (2005). It is worth pointing out that, apart from the transposon hypothesis, there was no reason whatsoever to suspect that transposons similar to RAG should exist.
Annotated Bibliography in Chronological Order
Burnet, F. M. (1971). "Self-recognition in colonial marine forms and flowering plants in relation to the evolution of immunity." Nature 232(5308): 230-235. (PubMed | DOI | Journal | Google Scholar)
The author of this article, Sir Frank MacFarlane Burnet, won the 1960 Nobel Prize in Medicine for the discovery of acquired immunological tolerance (http://nobelprize.org/medicine/laureates/1960/). He shared the award with Peter Medawar. Burnet wrote widely on the origin and evolution of the adaptive immune system in the 1960's and 1970's. Although he wrote before the molecular mechanism of recombination was understood he made many perceptive suggestions and is still commonly cited.
The 1971 Nature article is an early example of using comparative immunology in the explicit context of proposing and testing evolutionary hypotheses for the origin of the adaptive immune system. Burnet's specific point is that the ability to distinguish self from non-self is useful not just for fending off disease, but also for other purposes, e.g. detecting abnormal cancer cells, battling competing colonies in colonial tunicates, and preventing self-fertilization in flowering plants. The molecular mechanisms of recognition, such as complementary protein receptors on the cell surface, are not particularly difficult to understand. He argues that a basic self/nonself-recognition system could have provided a starting point for the evolution of the much more complex system of receptors used in the vertebrate adaptive immune system.
Burnet (1971) is cited repeatedly in the literature as outlining the subsequent research program on the origin of adaptive immunity. An example of one of Burnet's forward-looking statements:
"Much more extensive comparative studies are called for and in due course analysis of the results should allow a clear evolutionary history to emerge. Whatever form that history eventually takes we can be certain that gene duplication (gene expansion) plays a major part, and that progressive specialization of cell function and phenotypic restriction will be as conspicuous as it is in all other organs and functions." (p. 232)Citation of Burnet (1971) is sometimes accompanied by the following quote, "Comparative studies which are not made meaningful by use of evolutionary hypotheses are bound to be sterile." This is misattributed to the 1971 article; instead, it is actually the last sentence in a 1973 Nature article by Burnet ("Multiple Polymorphism in Relation to Histocompatibility Antigens," Nature 245(5425), pp. 359-361).
Marchalonis, J. J. (1976). Immunity in Evolution. Cambridge, Mass., Harvard University Press. (Library | Amazon | Google Print)
Early synthesis of knowledge on the evolution of the immune system, published in 1976. The forward, by Frank MacFarlane Burnet, suggests that the book might emerge as "the first definitive statement of the evolutionary origins and development of vertebrate immunity" (p. xx). The author, John Marchalonis, went on to be one of the leaders in the field of immune system evolution (see his articles in this bibliography through 2006).
Chapter 1 briefly reviews the history of evolutionary immunology, which goes back to at least to Metchnikoff in late 1800's. One of the major points that emerges from comparative immunology is that the defining features of mammalian immunity are shared to different degrees with relatives on the phylogenetic tree.
In the conclusion (p. 263), Marchalonis states, "many puzzling developments in human and mammalian immunity are clarified when analyzed in an evolutionary context." His final sentence makes the point that the study of evolution has practical benefit: "The study of the phylogeny of immunity still retains the excitement of new discoveries and contributes directly toward the applied questions of immunology."
Sakano, H., Hüppi, K., Heinrich, G. and Tonegawa, S. (1979). "Sequences at the somatic recombination sites of immunoglobulin light-chain genes." Nature 280(6): 288-294. (PubMed | DOI | Journal | Google Scholar)
Famous article (within the field) that proposed the transposon hypothesis for the origin of the V(D)J recombination system of adaptive immunity. This speculation was based upon the paper's report on the role that RSSs play in the mechanism of V(D)J recombination, a discovery fundamental to modern scientific understanding of the rearrangement mechanism."Evolution of light-chain genesAs of summer 2005, Sakano et al. (1979) had received an extraordinary 735 citations in the scientific literature, according to the Science Citation Index.
Thus, the V-J joining may well be a reversal of an ancient accidental insertion of an IS-like [IS = insertion sequence] DNA element...."
"Fig. 6 A hypothetical scheme for the evolution of the K-light-chain genes." (p. 294)
Manning, M. J., ed. (1980). Phylogeny of Immunological Memory. Developments in Immunology. Amsterdam, Elsevier/North-Holland Biomedical Press. (Library | Amazon | Google Print)
Thirty-one chapters by various authors on comparative immunology; Proceedings of the International Symposium on Immunological Memory held at the American Society of Zoologists Meeting in Tampa, Florida, USA, 28-30 December, 1979.
Klein, J. (1986). "Evolution of Mhc." Natural History of the Major Histocompatibility Complex. New York, John Wiley & Sons: 715-762. (Library | Google Print)
This textbook provides an overview of the Major Histocompatibility Complex (MHC), a system of proteins that help distinguish self from non-self. The final chapter, chapter 10, is entitled "Evolution" and gives a review of self-recognition in various animals.
The chapter begins (p. 716):"I believe it was Dobzhansky who said that in biology nothing makes sense except in the light of evolution. This dictum will undoubtedly apply also to the Mhc. I doubt very much if we will ever fully understand the true function of the Mhc (what purpose does the Mhc serve?), the presence of the class Ib genes in the Mhc region (why do they persist?), or the origin and purpose of the Mhc polymorphism, without exploring the source from which the Mhc sprang.Despite the conservative tone of this introduction, the technical discussion of MHC evolution continues for 45 pages.
I have used the future tense in this last sentence because the study of Mhc evolution has hardly begun. Although more and more papers are being published in which the authors use the term evolution to explain this or that observation, it is significant that at the time of writing this book no paper has been published in which the Mhc DNA of a nonmammalian species has been cloned. The following pages can testify how little we know about Mhc evolution. Most of the harvesters in molecular biology have moved on to reap the next field that has ripened and have no time to finish harvesting where they began. For this reason, progress will probably be slow, particularly in these times of profit-making science. The study of evolution makes no promises as far as eliminating cancer or even producing vaccine against AIDS is concerned, and it is not easy to justify in a grant application. True, one can still make headlines by cloning bits of DNA from some extinct animal, but this probably will not last very long. Sooner or later somebody will want to know what the cloning tells us about the animals, or even whether this information is worth knowing in the first place.
The study of Mhc evolution will probably be a slow process, which will not be marked out by spectacular successess. Yet, if we want to comprehend the Mhc, we must conquer this last frontier." (p. 716)
Kelsoe, G. and Schulze, D. H., eds. (1987). Evolution and Vertebrate Immunity: The Antigen-Receptor and MHC Gene Families. Austin, University of Texas Press. (Library | Amazon | Google Print)
Twenty-six articles on the evolution of the immune system (71 authors). Very technical.
Síma, P. and Vetvicka, V. (1990). Evolution of Immune Reactions. Boca Raton, CRC Press. (Library | Amazon | Google Print)
Sima & Vetvicka (Preface, p. iv), begin by saying, "The purpose of this book is to reconstruct the history and evolutionary pathways of immunity among the various forms of life." (p. iv) This book is a survey of the field and all the relevant lineages. It is quite technical although the introduction and conclusion are more accessible."We presume that, in the next few years, the fields of comparative and evolutionary immunology will provide, not only inspiration for further investigations in biomedicine, but also a number of practical applicable results." (p. vii)
"[I]f comparative immunology is to become an exact science, it must respect the laws and knowledge not only of experimental immunology, but also those of the evolutionary sciences." (p. vii)
"From among the vertebrate taxa, we have selected only the first three classes, the Agnatha, Chondrichthyes, and Ostichthyes, in order to show where and how the morpho-functional basis of the truly adaptive immunity of the endothermic tetrapods gradually evolved." (p. v)
"In spite of the relative youthfulness of comparative immunology as a new vigorous branch of modern immunology, an impressive amount of data in this field has been amassed in many laboratories all over the world. However, our knowledge of the history of immunity is still limited. A precise understanding of how the key steps in immune evolution were achieved needs further sophisticated study and comparative research work. Many gaps still need to be filled in. Nevertheless the overall picture of the evolution of the defense system in the animal kingdom, including that of immunity, is now sufficiently well known to provide an integral overview." (p. 229)
"Evolutionary immunology is by now a continually expanding discipline that promises to provide, in the nearest future, not only new fruitful ideas and new knowledge which will be fully utilized in practice, but also a clue to the principles controlling the defense mechanisms." (p. 230)
Warr, G. W. and Cohen, N., eds. (1991). Phylogenesis of Immune Functions. Boca Raton, CRC Press. (Library | Amazon | Google Print)
Another technical work dealing with immune systems of both vertebrates and invertebrates. The introduction (p. iii) begins by citing Metchnikoff (1882). Page iii continues:"[I]t can be argued that for a biologist to ignore the evolutionary history either of an organism or a complex system is as much an impediment to comprehending fully that organism or system as it is for a student of society to ignore the history of mankind." (p. iii)In chapter 9, "Evolutionary Origins of Immunoglobulin Genes," (pp. 171-189), Litman et al. write:
"[W]e have concentrated on areas in which we believe that recent exciting developments have led to new insights and understanding of immunity and its evolution in diverse species." (p. iii)"While the progressive phylogenetic development of immunoglobulin complexity is well documented, until recently little structural information at either the protein or the gene levels has been available with which to evaluate the mechanisms of evolution involved." (p. 173)In chapter 14, "Mechanisms of Molecular Evolution in the Immunoglobulin Superfamily," (pp. 295-316), Warr & Dover write:"Duplications, or much higher order multiplications, are obvious in the Ig family" (p. 305).Figure 4 of this article gives a "Hypothetical scheme for the evolution of Ig genes" (p. 304). The model is pre-transposon hypothesis.
Dreyfus, D. H. (1992). "Evidence suggesting an evolutionary relationship between transposable elements and immune system recombination sequences." Molecular Immunology 29(6): 807-810. (PubMed | DOI | Journal | Google Scholar)
This paper reports sequence similarities between V(D)J recombination signal sequences, and the termini of Tcl-like transposable sequences found in invertebrates. The authors note that this supports Sakano et al.'s 1979 hypothesis:"Sakano et al. (1979) have previously proposed that the vertebrate somatic recombination pathway evolved via the insertion of a transposable element into an ancestral gene encoding an antigen binding molecule, thus accounting for the simultaneous appearance of separated gene segments and a recombination mechanism for their reassembly." (p. 807)
"These similarities suggest that the Tcl transposition pathway may share common sequence-specific binding factors with the immunoglobulin somatic recombination pathway." (p. 807)
Síma, P. and Vetvicka, V. (1992). "Evolution of Immune Accessory Functions." Immune System Accessory Cells. Edited by L. Fornusek and P. Síma. Boca Raton, CRC Press: 1-55. (Library | Amazon | Google Print)
Long review of immune system cells across vertebrates and invertebrates. Cites Metchnikoff (1892) on p. 5. Figure 2 (p. 37) depicts the progressive emergence of various aspects of the immune system in the phylogeny of animals."The aim of this overview is to attempt to compare the evolutionary emergence of accessory cellular function and the role of various defense cell types in defense reactions in major natural assemblages of metazoan species." (p. 2)The article begins with a quote from Burnet (1973) in Nature: "Comparative studies which are not made meaningful by use of evolutionary hypotheses are bound to be sterile."
Bartl, S., Baltimore, D. and Weissman, I. L. (1994). "Molecular evolution of the vertebrate immune system." Proceedings of the National Academy of Sciences 91(23): 10769-10770. (PubMed | Journal | JSTOR | Google Scholar)
This is a short (2 pages) review article. In his 1996 book Darwin's Black Box, Behe critiqued it for vagueness, but even in 1996 there were longer review articles available (e.g. Thompson 1995), and despite the shortness, the basic ideas proposed in this article and elsewhere have been dramatically confirmed in the subsequent decade, as detailed in our 2006 Nature Immunology essay.
The last sentences of Bartl et al. (1994):"It is possible that the ancestors of RAG genes may have been horizontally transferred into a metazoan lineage at some relatively recent point in evolution. The newly introduced RAG genes may have acted, most likely with other proteins, on preexisting recombination signals (which consists of conserved heptamer and nonamer sequences) that may have been present for some other function or by random chance. In that view, the signal sequences captured the ancestors of present day TCR and immunoglobulin gene segments. Such a scenario is highly speculative but if true would imply a startling role for horizontal information transfer as the pivotal event in the evolution of vertebrate immunity." (p. 10770)
Beck, G., Cooper, E. L., Habicht, G. S. and Marchalonis, J. J., eds. (1994). Primordial Immunity: Foundations for the Vertebrate Immune System. New York, The New York Academy of Sciences. (Library | PubMed | Publisher | Amazon | Google Print)
This volume on comparative and evolutionary immunology resulted from a May 3-5, 1993 conference held in Woods Hole entitled "Primordial Immunity: Foundations for the Vertebrate Immune System." It contains 24 articles and 18 poster summaries, mostly examining the immune systems of various phylogenetically basal organisms. Three of the articles focusing on the origin of adaptive immunity are included in the articles list.
In the Preface to Primordial Immunity, editor Gail Habicht writes,"We know -- but we need to be convincing to others -- that studies of primordial immunity have tremendous importance:for the clues they give us to higher systems;The value of maintaining the biodiversity of our planet should be obvious: The gene pool possessed by even the lowliest creatures has the potential for enormous benefit to mankind." (p. xi, emphasis added)
for their unique strategies -- some of which may be exploitable for human benefit -- not just in medicine but in agriculture where biological pest control is a desirable alternative to toxic pesticides;
but, most importantly, for their own sake.
Hoffman, J. A., Janeway, C. A. and Natori, S., eds. (1994). Phylogenetic Perspectives in Immunity: The Insect Host Defense. Austin, R. G. Landes Company. (Library | Amazon | Google Print)
Fifteen chapters on the immune systems of basal vertebrates and arthropods in an evolutionary perspective. Three of the most relevant chapters are:
Chapter 6, by Iwanga et al., "Clotting Cascade in the Immune Response of Horseshoe Crab" (pp. 79-96).
Chapter 11, by Eric H. Davidson, "Stepwise Evolution of Major Functional Systems in Vertebrates, Including the Immune System" (pp. 133-142).
Chapter 12, by Alister Dodds, "Molecular and Phylogenetic Aspects of the Complement System" (pp. 143-155).
Marchalonis, J. J. and Schluter, S. F. (1994). "Development of an Immune System." Primordial Immunity: Foundations for the Vertebrate Immune System. Edited by G. Beck, E. L. Cooper, G. S. Habicht and J. J. Marchalonis. New York, New York Academy of Sciences. 712: 1-11. (Library | PubMed | Publisher | Google Print)
Overview of the distribution and origin of major pieces of the immune system, and some discussion of the selective pressures that may have been in play. The lead author, Marchalonis, has been working on the evolutionary origin of the immune system since the 1970's.
The introductory paragraph highlights some of the practical benefits of understanding the evolution of the immune system."There has always been considerable interest in understanding the evolutionary origins of the immune system,1-6 and this quest has recently been given impetus by the application of advanced methods in recombinant DNA technology7-14 and immunochemistry.15-18 An understanding of the genetic mechanisms underlying the capacity of vertebrates to respond to a potentially enormous (greater than 107) set of antigenic markers associated with pathogens or cancers that might never have been presented to the animal during its evolutionary development is of general theoretical importance for the understanding of anticipatory mechanisms19 and of practical value in modulating the immune response in autoimmunity or amplifying it in cases of immunodeficiency."The article is one of three from the special ANYAS volume Primordial Immunity which were included in thebibliography as particularly relevant to the origin of adaptive immunity. The volume contains dozens of other papers, however, so it is included in the books list as well.
Marchalonis, J. J., Hohman, V. S., Kaymaz, H., Schluter, S. F. and Edmundson, A. B. (1994). "Cell Surface Recognition and the Immunoglobulin Superfamily." Primordial Immunity: Foundations for the Vertebrate Immune System. Edited by G. Beck, E. L. Cooper, G. S. Habicht and J. J. Marchalonis. New York, New York Academy of Sciences. 712: 20-33. (Library | PubMed | Publisher | Google Print)
Review of the origin and dramatic diversification of the various immunoglobulins (Igs)."Data obtained recently on gene sequences of Igs of sharks, the ancestors of which diverged from those of mammals more than 400 million years ago, allow us to make detailed comparisons regarding the homologies among Ig V and C domains in evolution." (p. 21)
"We will analyze putative evolutionary relationships among canonical Igs and members of the Ig superfamily using highly conserved sequences from light and heavy chains of primitive vertebrates (e.g., the sandbar shark) as prototypes to ascertain similarities between Ig-related molecules of vertebrates and invertebrates." (p. 32)
Ohno, S. (1994). "MHC Evolution and Development of a Recognition System." Primordial Immunity: Foundations for the Vertebrate Immune System. Edited by G. Beck, E. L. Cooper, G. S. Habicht and J. J. Marchalonis. New York, New York Academy of Sciences. 712: 13-19. (Library | PubMed | Publisher | Google Print)
Argues for a relatively late origin of MHC (self-recognition capability), after the origin of adaptive immunity. This bucks the dominant view and the article explicitly takes a "devil's advocate" approach.
Stewart, J. (1994). The Primordial VRM System and the Evolution of Vertebrate Immunity. Austin, R. G. Landes. (Library | Amazon | Google Print)
This book is on the origin of the VRM system (VRM = Variable Region Molecules, see p. 9).
The evolutionary origin of various non-VRM immunity is covered on pp. 10-14 as background."[T]he early history of immunology was marked by an outstanding contribution which is a fine model of the methodology capable of rendering an evolutionary approach fruitful. The Russian Metchnikoff was exceptional, not only among the immunologists of his day but (unfortunately) among those of succeeding generations as well, in that his background was neither that of a chemist nor of a medical clinician, but truly that of a theoretically informed biologist." (p. 10)The book emphasizes duplication and exaptation (e.g., see exaptation reference on p. 10: "redeployment" of single-cell feeding ability ("phagocytosis") to immune system function in multicellular animals), but it does not discuss the transposon hypothesis, even though page 45 seems to be crying out for it. However, the hypothesis only seems to have begun a resurgence in about 1994.
Stewart (1994) is cited by Behe on p. 282, note 7. Stewart is also the author of: Stewart, J. (1992) Immunoglobulins did not arise in evolution to fight infection. Immunology Today 13, 396-399.
Vetvicka, V., Síma, P., Cooper, E. L., Bilej, M. and Roch, P. (1994). Immunology of Annelids. Boca Raton, CRC Press. (Library | Amazon | Google Print)
Ten chapters on the immunology of annelids (earthworms and relatives). This may seem obscure, but the importance of annelids to evolutionary immunology is explained in Chapter 1, "Comparative Immunology: The Value of Annelids.""[A]ll invertebrates, including annelids, have figured prominently in establishing the field of immunology." (p. 6)
"As long as there are invertebrates like earthworms, oysters, or honey bees to be protected, and others like tapeworms, slugs, and mosquitoes to be destroyed, there will be a utilitarian justification for studying invertebrate paleontology and whatever is equivalent in them to what we study in vertebrates as immunology.... Most contributors are interested mainly in what, if any, light such studies of invertebrates can throw on the evolution of the processes of inflammation and immunity as we see them in humans and the experimental mammals of the laboratory. Since Darwin, much of the 'fun' of biological research has been to interpret what directly interests one in evolutionary terms." (pp. 5-6, quoting Burnet 1971 in Nature)
Thompson, C. B. (1995). "New insights into V(D)J recombination and its role in the evolution of the immune system." Immunity 3(5): 531-539. (PubMed | DOI | Journal | Google Scholar)
A long review article summarizing the early results supporting the transposon model for the origin of adaptive immunity."Current evidence suggests that the ability to mount an antigen-specific immune response arose over a relatively short period of time coincident with the development of the first jawed vertebrates. This has raised the question of how a mechanism as complex as V(D)J recombination arose over such a short evolutionary period. Recent advances in V(D)J recombination suggest a resolution to this issue and allow us to reexamine the role of antigen specificity in shaping the evolution of the immune system." (p. 531)
"These observations are already renewing speculation concerning the origins of RAG1 and RAG2." (p. 532).
"Conclusion
Recent advances in our understanding of V(D)J recombination and the distribution of immunoglobulin and TCR genes in vertebrate evolution give support to the hypothesis that the V(D)J recombination arose abruptly during early vertebrate evolution. Current evidence suggests that V(D)J recombination is directed by elements that could have been derived from a transposon. [...] Colonization of the genome of an ancestral vertebrate by a transposon represents a fundamental failure of the defense systems of the organism. It is ironic that the participation of such an element in the evolution of antigen-specific immunity may have played an important role in the evolutionary success of vertebrates." (p. 538)
Bernstein, R. M., Schluter, S. F., Bernstein, H. and Marchalonis, J. J. (1996). "Primordial emergence of the recombination activating gene 1 (RAG1): Sequence of the complete shark gene indicates homology to microbial integrases." Proceedings of the National Academy of Sciences 93(18): 9454-9459. (PubMed | Journal | JSTOR | Google Scholar)
This study compared RAG1 and RAG2 in sharks and mammals, delineating the highly conserved regions. These conserved regions were found to exhibit homology to bacterial recombinases."Homology domains identified within shark RAG I prompted sequence comparison analyses that suggested similarity of the RAG I and II genes, respectively, to the integrase family genes and integration host factor genes of the bacterial site-specific recombination system. Thus, the apparent explosive evolution (or "big bang") of the ancestral immune system may have been initiated by a transfer of microbial site-specific recombinases." (p. 9454)Figure 5 contains a general phylogenetic tree of the prokaryotic relatives of RAG1, and depicts the hypothesized horizontal transfer event.
van Gent, D. C., Mizuuchi, K. and Gellert, M. (1996). "Similarities between initiation of V(D)J recombination and retroviral integration." Science 271(5255): 1592-1594. (PubMed | Journal | JSTOR | Google Scholar)
This study reports close similarities between the mechanism of the formation of DNA hairpins during the cutting and assembly of antibody genes, and the processes that occur when transposons and retroviruses insert into DNA."These findings provide support for the speculation that the antigen receptor genes, and the RAG1 and RAG2 proteins that mediate their rearrangement, may have evolved from an ancestral transposon (18). The presence of RSSs facing in opposite directions (the most usual arrangement in the antigen receptor loci) is similar to the inverted repeat architecture of many transposon ends, and the RAG proteins could have developed from genes encoded by the former transposon."Ref. 18 is Thompson (1995).
Hughes, A. L. and Yeager, M. (1997). "Molecular evolution of the vertebrate immune system." BioEssays 19(9): 777-786. (PubMed | Google Scholar)
Review of the evolution of several key pieces of the immune system (e.g., C3 and the other complement proteins, MHC, RAG & immunoglobulins, etc.), with a focus on the evolutionary mechanisms involved, e.g. various forms of gene duplication vs. block duplication.
Several of the key questions asked here in 1997 have been answered. The discussion of the origin of RAGs is quoted to show what was known then, and what research questions were being asked."Some major unanswered questionsThe cited references in this passage include the familiar Bernstein et al. (1996):
(1) The origin of the immunoglobulin superfamily C1 type of domain
[...]
(2) Block duplication vs. functional gene clustering
[...]
(3) Conserved proteins involved in vertebrate adaptive immunity
One of the unique features of the vertebrate immune system is the rearrangement of gene segments to produce expressed Ig and TCR genes. Because these gene segments are short and have evolved rapidly, it may be very difficult to reconstruct their early evolutionary history. It may be possible, however, to obtain clues regarding the early history of Ig and TCR by studying the phylogeny of conserved proteins playing key roles in the process of rearrangement. These include the products of two genes called RAG-1 and RAG-2, which are linked in mammals. Homologues of RAG-1 have been sequenced from mammals, amphibians, bony fishes and shark(63). The RAG-1 protein shows some evidence of homology to the bacterial site-specific recombinases Fim B and Fim E(63), while RAG-2 shows homology to the bacterial recombinases Hin A(63). A phylogenetic analysis (Fig. 5), based on the region of homology between RAG-1, FimB and Fim E, illustrates this relationship. RAG-1 shows homology to eukaryotic proteins involved in such processes as excision repair (yeast RAG16(64) and regulation of gene expression (mouse rpt-1r(65) and to a human acid finger protein (66)). Interestingly, in the phylogenetic tree, RAG-1 clusters closer to the bacterial proteins than it does to any of these eukaryotic proteins (Fig. 5).
The homologies between RAG-1 and RAG-2 and bacterial proteins led Bernstein et al.(63) to suggest that somatic recombination arose in vertebrates as a result of horizontal transfer of a mictobial [sic] recombinase gene. The mere fact of homology between RAG-1 and RAG-2 and bacterial genes, however, is not evidence of horizontal gene transfer, since many vertebrate genes have prokaryotic homologues. [...] Nonetheless, Bernstein et al.(63) are correct in pointing out the potential importance of understanding the origin of this gene, and others involved in the recombination process, for gaining insights into the origin of adaptive immunity." (p. 785)[References]
63 Bernstein, R., Schluter, S.F., Bernstein, H. and Marchalonis, J.J. (1996). Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc. Natl. Acad. Sci. USA 93, 9454-9459.
64 Bang, D.d., Verhage, R., Goosen, N., Brouwer, J. and van de Putte, P. (1992). Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res. 20, 3924-3931.
65 Patarca, R. et al. (1988). rpt-1, an intracellular protein from helper/inducer T cells that regulates gene express of interleukin 2 receptor and human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 85, 2733-2737.
66 Chu, T.W., Capossela, A., Coleman, R., Goel, V.L., Nallur, G. and Gruen, J.R. (1995). Cloning of a new 'finger' protein gene (ZNF173) within the class I region of the human MHC. Genomics 29, 229-239.
67 Hughes, A.L. (1994). Evolution of the ATP-binding cassette transmembrane transporters of vertebrates. Mol. Biol. Evol. 11, 899-910.
Ji, X., Azumi, K., Sasaki, M. and Nonaka, M. (1997). "Ancient origin of the complement lectin pathway revealed by molecular cloning of mannan binding protein-associated serine protease from a urochordate, the Japanese ascidian, Halocynthia roretzi." Proceedings of the National Academy of Sciences 94(12): 6340-6345. (PubMed | Journal | Google Scholar)
Research paper reporting that key innate immune system genes have been discovered in tunicates, invertebrates related to vertebrates (also known as ascidians or sea-squirts)."The major developments of the complement system, therefore, seem to have occurred in two stages; first, with the emergence in cyclostomes of the alternative pathway to establish a means of amplification, and second, after the divergence of cyclostomes but before the divergence of cartilaginous fish, the emergence of the classical and lytic pathways. This hypothesis would not only explain the evolution of the complement system, but also may help to reevaluate the activation mechanism of the mammalian alternative pathway." (pp. 6344-6345)
Lewis, S. M. and Wu, G. E. (1997). "The origins of V(D)J recombination." Cell 88(2): 159-162. (PubMed | DOI | Journal | Google Scholar)
Lewis and Wu have authored several reviews on the evolutionary origin of the adaptive immune system. This 1997 review in Cell summarizes the state of the question after several then-recent discoveries. The introductory section, "The Origins of V(D)J Recombination," discusses why the evolutionary question was attracting renewed interest:"Recently V(D)J recombination has been partially reconstituted in vitro (reviewed in Gellert, 1996), and as a result, has been the subject of intense research. A side effect of these efforts has been renewed speculation regarding evolutionary origins. New information bears upon the possibility that this unusual recombination system could have been a transplant from the procaryotic world (Difilippantonio et al., 1996; Spanopoulou et al., 1996, van Gent et al., 1996a)." (p. 159)This article reviews "The Case for a Transposon Progenitor" of RAG (section heading, p. 161), e.g.:"Given that the architecture of a joining signal is transposon-like, this similarity between chemical mechanisms would seem to add weight to the idea that the V(D)J recombination system originally carried out transpositional rearrangements. The transposon theory has in fact been suggested, for various reasons, a number of times over the years." (p. 162)On the other hand, several similarities were missing in 1997:"It is nonetheless worth noting that in spite of various similarities, there are fundamental differences between transposition and V(D)J recombination. For one, there are no reports of a transposase (mutant or otherwise) that is able to mediate site-specific inversion. Conversely, it has never been demonstrated that V(D)J recombination can cause the integration of one piece of DNA into another." (p. 162)Both of these features were soon discovered in lab experiments in following years.[References]
Difilippantonio, M.J., McMahan, C.J., Eastman, Q.M., Spanopoulou, E., and Schatz, D.G. (1996). Cell 87, 253-262.
Gellert, M. (1996). Genes to Cells 1, 269-276.
Lewis, S.M. (1994). Adv. Immunol. 56, 27-150.
Spanopoulou E., Zaitseva, F., Wang, F.-H., Santagata, S., Baltimore, D., and Panaoytou, G. (1996). Cell 87, 263-276.
van Gent, D.C., Mizuuchi, D., and Gellert, M. (1996a). Science 271, 1592-1594.
van Gent, D.C., Ramsden, D.A., and Gellert, M. (1996b). Cell 85, 107-113.
Schluter, S. F., Bernstein, R. M. and Marchalonis, J. J. (1997). "Molecular origins and evolution of immunoglobulin heavy-chain genes of jawed vertebrates." Immunology Today 18(11): 543-549. (PubMed | DOI | Journal | Google Scholar)
A review of the origins of rearranging immunoglobulins in the light of cartilagenous fish, the most basal vertebrate group possessing classic adaptive immunity. The introduction clearly states the kinds of questions that an evolutionary model must answer."A model for the evolution of the immunoglobulins (Igs) must explain two separate but closely related aspects. The first is the genetic mechanisms by which the diversity of the expressed effector molecules is generated1. This would encompass both the enzymatic mechanism of recombination and the genomic organization of the various elements of the Ig genes. The second is the functional duality of the receptor proteins whereby the variable (V) regions form the antigen-binding sites and the effector activities are mediated through the constant (C) regions2. This article will focus on the latter aspect, particularly the molecular evolutionary origins of the gene segments encoding Ig heavy (H) chains." (p. 543)A model for the origin of the various immunoglobulins is constructed based on the phylogenies reviewed in the paper:"A model for the evolution of the VH genes
From the evidence discussed above, a model for the evolutionary origins of VH and Ig isotypes is presented in Fig. 4. As originally proposed, the V and C domains arose by duplication of a primordial Ig domain3,13. The C domains duplicated and evolved independently to form the primordial prototype H-chain isotype." (p. 548)
Agrawal, A., Eastman, Q. M. and Schatz, D. G. (1998). "Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system." Nature 394(6695): 744-751. (PubMed | DOI | Journal | Google Scholar)
This study reports a major research finding that supported the transposon hypothesis for the origin of adaptive immunity. The authors found that the rearrangment-activating genes, RAG1+RAG2, could still perform both the excision and the insertion reactions, just like a free-living transposon.
Figure 7 is a nice color graphic of the transposon hypothesis."Our results are evidence in favour of the theory that a vital event in the evolution of the antigen-specific immune system was the insertion of a 'RAG transposon' into the germ line of a vertebrate ancestor14,41." (p. 750)
[References]
14. Thompson, C. B. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity 3, 531-539 (1995).
15. Lewis, S. M. & Wu, G. E. The origins of V(D)J recombination. Cell 88, 159-162 (1997).
41. Sakano, H., Hüppi, K., Heinrich, G. & Tonegawa, S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280, 288-294 (1979).
Plasterk, R. (1998). "Ragtime jumping." Nature 394(6695): 718-719. (PubMed | DOI | Journal | Google Scholar)
Summary of the Nature article Agrawal et al. (1998). While short and nontechnical, it is relevant to some of Behe's "search criteria" as he laid out during his sworn deposition for the Kitzmiller case. During his deposition in May 2005, Behe was quizzed about some of his claims about the immune system, and he was asked how he even knew what was in the literature since he wasn't familiar with the peer-reviewed articles that were presented then. Behe's answers (Behe deposition, pp. 231-233) were basically (1) he relies on other people to email him articles that meet his challenges, (2) articles on the evolution of the immune system would be big news, so he watches out for news articles in major scientific publications, and review articles in review journals and other scientific literature, and (3) he watches out for articles in popular scientific outlets such as the New York Times or Scientific American.
Plasterk's 1998 article in Nature is an example of case #2, which Behe missed. Many of the other articles in this bibliography are also review articles; one is even found in Annual Review of Immunology (Litman et al. 1999), and two are in Immunological Reviews (Cannon et al. 2004, Jones et al. 2004). Although not included in this bibliography, two 1996 articles in Scientific American on the evolution of the immune system are examples of case #3.
The Plasterk article begins:
"Transposons are generally considered the ultimate forms of selfish DNA -- a single gene (or sometimes a set of two or three genes) that spreads simply because it ensures its own replication. Indeed, the ability of transposons to encode one or more proteins that selectively replicate the transposon is enough to explain their existence. But selfish elements may also end up doing something useful for their host, and it has often been speculated that transposon jumping may have generated gene arrangements that opened new avenues in evolution. Examples of this are rare, but a spectacular case has now been discovered. On page 744 of this issue, David Schatz and his colleagues report that we owe the repertoire of our immune system to one transposon insertion, which occurred 450 million years ago in an ancestor of the jawed vertebrates. Vertebrates seem to have tamed this ancient transposon for generation of the immune repertoire, and the authors show that the RAG1 and RAG2 proteins (which mediate V(D)J joining) can still catalyse a full transposition reaction. A similar result has been independently obtained by Martin Gellert and co-workers2, and is reported in tomorrow’s issue of Cell."
[References]
1. Agrawal, A., Eastman, Q. M. & Schatz, D. G. Nature 394, 744-751 (1998).
2. Hiom, K., Melek, M. & Gellert, M. Cell 94, 463-470 (1998).
Vetvicka, V. and Síma, P. (1998). Evolutionary Mechanisms of Defense Reactions. Basel, Birkhäuser Verlag. (Library | Amazon | Google Print)
Vetvicka & Sima systematically review the immune systems of animals, starting with the earliest branches on the animal phylogenetic (sponges), through the various simple and complex invertebrates, the chordate relatives of vertebrates, the early diverging vertebrates such as the lampreys and hagfish, and ending with the "higher" vertebrates and their adaptive immune systems. Highly technical, focuses on immune system organs in addition to the cellular/molecular level. The primary themes are that: (1) immune system evolution must be considered in the context of ecological and morphological change in mammal groups; (2) there is a large degree of diversity in so-called "primitive" organisms, and there are many ways organisms survive without the "required" parts of vertebrate immunity."[B]asic research in comparative and evolutionary immunology has substantially contributed in many ways to the exciting progress that immunology has made in recent decades." (p. 187)
Lewis, S. M. (1999). "Evolution of Immunoglobulin and T-Cell Receptor Gene Assembly." Annals of the New York Academy of Sciences 870: 58-67. (PubMed | Journal | Google Scholar)
Another detailed review of the RAG transposon hypothesis by Susanna Lewis. It is a clear example of the increasing confidence that researchers in the field have about the transposon hypothesis. E.g., from the introductory section, entitled, "A transposon origin for V(D)J recombination":"How such a singular mechanism for somatic cell differentiation arose has long been a puzzle.6 Although a multiplicity of site-specific recombination systems exist in bacteria, yeast, and protozoa, no other example apart from V(D)J recombination is known to operate in vertebrates. One speculation dating back to the time of the first molecular descriptions of immunoglobulin gene rearrangement is that the V(D)J recombination system may have evolved from a mobile element.7 Now, almost 20 years later, biochemical analyses of RAG1 and RAG2 have uncovered solid clues that this indeed may be the case." (p. 58)Figure 4 contains a summary depiction of the transposon hypothesis. Lewis (1999) is especially noteworthy because it contains an uncannily predictive suggestion in the section suggesting further research avenues:"THE NEXT STEPExactly this was discovered six years later, in 2005 (Kapitonov & Jurka, 2005). Another cousin was discovered in 2006 (Fugmann, S. D., Messier, C., Novack, L. A., Cameron, R. A. and Rast, J. P. (2006). "An ancient evolutionary origin of the Rag1/2 gene locus." Proc Natl Acad Sci U S A 103(10): 3728-3733)
It would be extremely useful if a contemporary version of the original RAG transposon could be identified. A distant cousin, with credentials, would greatly facilitate any attempts to reconstruct the lost history between the time the first RAG element took up residence in the vertebrate genome and the emergence of a developmental recombination system." (p. 65)[References]
1. Marchalonis, J.J., S.F. Schluter, R.M. Bernstein, S. Shen & A.B. Edmundson. 1998. Phylogenetic emergence and molecular evolution of the immunoglobulin family. Adv. Immunol. 70: 417-506.
2. Gellert, M. 1997. Recent advances in understanding V(D)J recombination. Adv. Immunol. 64: 39-64.
[...]
6. Lewis, S. & G. Wu. 1997. The origins of V(D)J recombination. Cell 88: 159-162.
7. Sakano, H., K. Hüppi, G. Heinrich & S. Tonegawa. 1979. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280: 288-294.
Litman, G. W., Anderson, M. K. and Rast, J. P. (1999). "Evolution of antigen binding receptors." Annual Review of Immunology 17: 109-147. (PubMed | DOI | Journal | Google Scholar)
This very long (38 pages) and technical review article cites 171 references, surveying all aspects of the origins of adaptive immunity, focusing on the ancestry and phylogeny of the receptors rather than the transposon hypothesis which is taken as confirmed."This review addresses issues related to the evolution of the complex multigene families of antigen binding receptors that function in adaptive immunity. [...] As of yet, homologous forms of antigen binding receptors have not been identified in jawless vertebrates; however, acquisition of large amounts of structural data for the antigen binding receptors that are found in a variety of jawed vertebrates has defined shared characteristics that provide unique insight into the distant origins of the rearranging gene systems and their relationships to both adaptive and innate recognition processes." (p. 109)
"Further clarification of the origins and interrelationships of these putative receptors will come through phylogenetic comparisons between rearranging and nonrearranging antigen binding receptors found in jawed vertebrates. Such analyses will also be significant in terms of designing strategies for identification of related systems in agnathans, lower chordates, hemichordates, and echinoderms. The recent discovery that RAG1 and RAG2 proteins together constitute a transposase, capable of excising a piece of DNA containing recombination signals from a donor site and inserting into a target DNA molecule, has enormous bearing on mechanisms whereby nonrearranging receptors diverged into the rearranging antigen binding receptor genes (171)." (p. 139)
[References]
171. Agrawal A, Eastman QM, Schatz DG. 1998. Transposition mediated by the V(D)J recombination proteins RAG1 and RAG2: implications for the origins of the antigen-specific immune system. Nature 394:744-51
Schatz, D. G. (1999). "Transposition mediated by RAG1 and RAG2 and the evolution of the adaptive immune system." Immunologic Research 19(2-3): 169-182. (PubMed | Google Scholar)
Technical review of the tranposon hypothesis. The abstract summarizes Schatz's main point:"The hypothesis that RAG1 and RAG2 arose in evolution as components of a transposable element has received dramatic support from our recent finding that the RAG proteins are a fully functional transposase in vitro."Noteworthy in the article is Schatz's description of Sakano et al. (1979) as advancing a "prophetic hypothesis":"When examined in their germline configuration, the RSSs flanking V and J constitute an inverted repeat, much like that found at the end of transposons, and this realization led to the prophetic hypothesis, in 1979, that insertion of a transposable element into an ancient receptor gene exon was responsible for the generation of split antigen receptor genes during evolution (5)." (p. 170)
[Reference cited]
5 Sakano H, Hüppi K, Heinrich G, Tonegawa S: Sequences at the somatic recominbation sites of immunoglobulin light-chain genes. Nature 1979;280:288-294.
Schluter, S. F., Bernstein, R. M., Bernstein, H. and Marchalonis, J. J. (1999). "'Big Bang' emergence of the combinatorial immune system." Developmental and Comparative Immunology 23(2): 107-111. (PubMed | DOI | Journal | Google Scholar)
Basic review of the transposon hypothesis. The title and various quotes are the kind of thing that are commonly quote-mined by creationists, e.g., "Thus, the Big Bang must have occurred in the extremely short time of emergence of jawed vertebrates from their ostracoderm ancestors." (p. 107) However, it is worth noting that "extremely short" means about 50 million years in this context. And, the whole point of the article is that the transpson hypothesis, where a prokaryotic transposon jumped into the vertebrate genome, is what explains the "big bang" in question.
For example:"We [1, 2, 20] and others [6, 8, 33, 34] have hypothesized that the event that catalyzed the explosive burst of the generation of combinatorial immunity (Fig. 1) was the horizontal transfer of genes from microbes and/or fungi enabling site-specific recombination of DNA. Because retroviruses have long been known to insert into mammalian chromosomes and defective pieces of retrotransposons have been found in lower deuterostomes [33, 34], sharks [35], turtles [36] and chickens [37], and retrotransposons are widely distributed among teleost fish [38], we tentatively suggested that retroviruses may have played a role in this transition. However, transposons capable of directly modifying DNA [39, 40] may also play a substantial role. It was first recognized by Thompson [6] that the gene cluster specifying RAG1 and RAG2 resembles a disassociated transposon, thus suggesting a transposon origin for these genes. Direct in vitro experimental support for this hypothesis was recently provided in two reports, Agrawal et al., [41] and Hiom et al.[42]. These groups showed that recombinant RAG1 and RAG2 proteins together could cleave a fragment of DNA flanked by RSS (recombinant signal sequence) sites and mediate its insertion into a plasmid, thus directly demonstrating that RAG proteins can express transposase activity." (p. 108)Figure 2 gives a depiction of the transposon hypothesis. More evidence for the transposon hypothesis is given on the next pages:"IHF [Integration Host Factor] binding sites in some transposons (e.g. TnA family) occur adjacent to transposase binding sites, suggesting that the RAGI/RAGII recombination system arose from an ancestral system in a transposon or retrotransposon." (pp. 109-110)
[References]
[1] Marchalonis JJ, Schluter SF, Bernstein RM, Edmundson AB. Phylogenetic emergence and molecular evolution of the immunoglobulin family. Adv. in Immunol. 1998; 70:417-506.
[2] Marchalonis JJ, Schluter SF. A stochastic model for the rapid emergence of specific vertebrate immunity incorporating horizontal transfer of systems enabling duplication and combinatorial diversification. J. Theo. Biol. 1998; 193:429-444.
[3] Marchalonis JJ, Schluter SF. On the relevance of invertebrate recognition and defense mechanisms to the emergence of the immune response of vertebrates. Scand. J. Immunol. 1990;32:13-20.
[4] Klein J. Homology between immune responses in vertebrates and invertebrates: does it exist? Scand. J. Immunol. 1997;46:558-564.
[5] Hughes AL, Yeager M. Molecular evolution of the vertebrate immune system. BioEssays 1997;19:777-786.
[6] Thompson CB. New Insights into V(D)J recombination and its role in the evolution of the immune system. Immunity 1995;3:531-539.
[7] Medzhitov R, Janeway CA. Innate Immunity: The virtues of a nonclonal system of recognition. Cell 1997;91:295-298
[8] Bartl S, Baltimore D, Weissman IL. Molecular evolution of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 1994;91:10769-10770.
[9] Habicht GS. Primordial immunity: foundations for the vertebrate immune system. In: Beck G, Cooper EL, Habicht GS, Marchalonis JJ, editors. Primordial Immunity: Foundations for the Vertebrate Immune System, vol. 712. New York: New York Academy of Sciences, 1994:ix-xi.
[41] Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 1998;394:744-751.
[42] Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 1998;94:463-470.
Du Pasquier, L. (2000). "The Phylogenetic Origin of Antigen-Specific Receptors." Origin and Evolution of the Vertebrate Immune System. Edited by L. Du Pasquier and G. W. Litman. Berlin, Springer. 248: 159-185. (Library | PubMed | Amazon | Google Print)
Highly technical review of the origin of the receptor genes, starting with Ig homologs in invertebrates, and then looking at the relationships between the various immune system genes within vertebrates."Fig. 10. Origin and evolution of Igsf members with V and C1 domains. A hypothesis taking into account the genetic linkages and the homologies presented in this chapter." (pp. 178-179)
"This survey reminds us that V domains are very ancient and exist in the most primitive Metazoa. [...] A model (Fig. 10) is presented accounting for many of the linkages and homologies described." (p. 182)
Du Pasquier, L. and Litman, G. W., eds. (2000). Origin and Evolution of the Vertebrate Immune System. Current Topics in Microbiology and Immunology. Berlin, Springer. (Library | Publisher | Amazon | Google Print)
This edited book in the Current Topics in Microbiology and Immunology series contains 14 articles by 33 contributors on the evolutionary origin of the immune system. It is a thorough survey of the evolution of the major parts of the immune system as of 2000. Most of the articles are very technical. Despite this, the Preface reads,"This book is by no means comprehensive and can be complemented by reading recent issues of Immunological Reviews (nos. 166, Immune systems of ectothermic vertebrates, and 167, Genomic organisation of the MHC: structure, origin, and function)." (p. vi)Also, from Chapter 1, Section 4, "The Evolution of Complex Systems":
"The quantity of information concerning vertebrate immunity from developmental and molecular perspectives is enormous, and offers an excellent opportunity to study the evolutionary behavior of complex molecular networks. As more sea urchin genes that are homologous to genes of the vertebrate immune system are isolated, it is becoming clear that the phylogenetic position of the echinoderms will truly enable the creation and testing of hypotheses of large scale immune system evolution." (Chapter 1, "New approaches Towards and Understanding of Deuterostome Immunity", p. 5)"Subsystems of a complex immune system may evolve independently from one another, and as a first step towards understanding system-wide evolution these points of disjunction must be characterized." (Chapter 1, "New approaches Towards and Understanding of Deuterostome Immunity", p. 12)
Hansen, J. D. and McBlane, J. F. (2000). "Recombination-Activating Genes, Transposition, and the Lymphoid-Specific Combinatorial Immune System: A Common Evolutionary Connection." Origin and Evolution of the Vertebrate Immune System. Edited by L. Du Pasquier and G. W. Litman. Berlin, Springer. 248: 111-135. (Library | PubMed | Amazon | Google Print)
This is one of three articles from the volume, "Origin and Evolution of the Vertebrate Immune System," which were included in the article list. The book itself is listed in the book list. The three articles were included in the article bibliography because they were particularly useful summaries of the evolution of key pieces of adaptive and innate immunity.
This article is a technical review of the evolution of V(D)J recombination and evolution. Section 4, "Origins of Rag1 and Rag2," contains the now-familiar review of the development and confirmation of the transposon hypothesis:"The genomic organization of the Ig and TCR loci themselves first hinted at a possible link between the V(D)J recombination mechanism and DNA transposition (Sakano et al. 1979). Detailed knowledge of the Rag genes and the biochemical activities of their products strengthened this comparison (Lewis and Wu 1997; Thompson 1995). Recent biochemical data sealed this functional link, strongly suggesting that the Rag genes appeared in the vertebrate genome about 450 MYA as passengers in a transposon. The evidence accumulated to date in favor of the Rag transposon comes from 4 main sources..." (p. 122)The lines of evidence presented should be familiar at this point; however, the authors highlight the importance of the fourth line of evidence, "DNA transposition by the Rag proteins":"The most dramatic parallel between DNA transposition and V(D)J recombination was shown recently by two groups (Agrawal et al. 1998; Hiom et al. 1998; reviewed in Plasterk 1998; Roth and Craig 1998). In these studies, the Rag proteins performed not only DNA cleavage at the RSS, but also transpositional insertion of the RSS-containing cleavage products into unrelated target DNA (Fig. 4)." (p. 123)The authors continue:"The structure of the antigen receptor loci is itself a compelling argument in favor of a transpositional origin for the immune system (Sakano et al. 1979), with both V(D)J recombination and DNA transposition involving a pair of inverted repeats at the ends of the recombining agent." (p. 123)On p. 125, Figure 5 shows the "Rag transposon model".The concluding section of the article reviews what has gone before: "We describe multiple lines of evidence above which suggest that the Rag genes were introduced into the vertebrate genome over half a billion years ago as part of an ancestral transposon." (p. 130)...and follows it up with a summary of the model.
Lewis, S. M. and Wu, G. E. (2000). "The old and the restless." Journal of Experimental Medicine 191(10): 1631-1636. (PubMed | DOI | Journal | Google Scholar)
Another review paper on the RAG transposon hypothesis from Lewis and Wu. Figure 2 contains a usefully clear figure depicting the hypothesis. The second paragraph sums up the development of the transposon hypothesis:"As recently as only a few years ago, any real information bearing on the actual genesis of the V(D)J recombination system seemed to be irretrievably lost. However, as more was learned about the molecular genetic and biochemical properties of the V(D)J recombination proteins, termed recombination activating gene (RAG)-1 and RAG-2, tantalizing suggestions of a transposon origin began to emerge. These clues included the following: first, the fact that the genes encoding RAG-1 and RAG-2, which are unrelated in sequence, are tightly linked, and as such share this property with genes that are known to undergo horizontal transfer (4). Second, the chemical mechanism of the recombination reaction, where DNA strand breakage and rejoining is accomplished through one step transesterification reactions, was like that of several well-described mobile elements (5). Finally, a surprising finding further indicated that RAG-1 and RAG-2 might have once been part of a transposon when two groups independently demonstrated that purified RAG-1 and RAG-2 proteins have a latent ability to carry out the transposition of DNA (6, 7)." (p. 1631)
"Thus, the unusual in vitro ability of RAG-1 and RAG-2 to do two quite different things, and in particular to transpose DNA, provided strong support for the original speculation that the V(D)J recombination system used to be a transposable element (3)." (p. 1631)
[References]
1. Marchalonis, J.J., S.F. Schluter, R.M. Bernstein, S. Shen, and A.B. Edmundson. 1998. Phylogenetic emergence and molecular evolution of the immunoglobulin family. Adv. Immunol. 70:417-506.
2. Litman, G.W., M.K. Anderson, and J.P. Rast. 1999. Evolution of antigen binding receptors. Annu. Rev. Immunol. 17:109-147.
3. Sakano, H., K. Huppi, G. Heinrich, and S. Tonegawa. 1979. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 280:288-294.
4. Oettinger, M.A., D.G. Schatz, C. Gorka, and D. Baltimore. 1990. Rag-1 and Rag-2, adjacent genes that synergistically activate V(D)J recombination. Science. 248:1517-1523.
5. van Gent, D.C., D. Mizuuchi, and M. Gellert. 1996. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 271:1592-1594.
6. Agrawal, A., Q.M. Eastman, and D.G. Schatz. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 394:744-751.
7. Hiom, K., M. Melek, and M. Gellert. 1998. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 94:463-470.
Nonaka, M. (2000). "Origin and evolution of the Complement System." Origin and Evolution of the Vertebrate Immune System. Edited by L. Du Pasquier and G. W. Litman. Berlin, Springer. 248: 37-50. (Library | PubMed | Amazon | Google Print)
Nonaka regularly authors or coauthors articles reviewing the evolutionary origin of the innate immune system. This article surveys the complement systems of vertebrates and invertebrates and focuses on the origin of the B and C2 proteins which help activate the complement system, and the MASP proteins, mammalian serine proteases which actually have homologs in groups as distant as ascidians."This chapter discusses recent progress in understanding the evolution of the complement system mainly in invertebrates and lower vertebrates." (p. 38)
"The hypothetical story of the origin and evolution of the complement system has been composed from data discussed above (Fig. 3). The birth of the complement system is located somewhere during evolution of invertebrate phyla, although the exact stage at which the complement system was established must still be clarified. The original complement system most probably resembled the LeP and AP of the mammalian complement system. Lectin-based recognition by MBL, followed by the activation of MASP, B, and then C3, covalent binding of C3 to foreign particles, and finally phagocytosis of C3-tagged foreign particles by phagocytes with C3 receptors on their surface seem to be the most probable constitution of the original complement system." (p. 46)
Opal, S. M. (2000). "Phylogenetic and functional relationships between coagulation and the innate immune response." Critical Care Medicine 28(9): S77-S80. (PubMed | Journal | Google Scholar)
This article reviews the evolutionary and physiological relationships between the blood-clotting cascade (coagulation) and the the inflammatory cascade which initiates innate immune responses."The simultaneous activation of the inflammatory response and the coagulation system after injury is a phylogenetically ancient, adaptive response that can be traced back to the early stages of eukaryotic evolution." (p. S77)
"Concluding Remarks. The coagulation system is integrally related to the innate immune response, and its activation and regulation are dependent on local and systemic immune responses. The simultaneous activation of clotting and the innate immune response is a phylogenetically ancient host response to tissue injury and is a primary survival strategy throughout vertebrate evolution." (p. S79)
Richards, M. H. and Nelson, J. L. (2000). "The Evolution of Vertebrate Antigen Receptors: A Phylogenetic Approach." Molecular Biology and Evolution 17(1): 146-155. (PubMed | Journal | Google Scholar)
This research article explores the phylogeny of somatically rearranging antigen receptors, seeking the most basal groups."It is now clear that indirect, MHC-restricted antigen recognition must be a derived characteristic of [alpha][beta] T cells, while [gamma][beta] T cells exhibit the primitive condition of direct antigen binding. This removes a major stumbling block in several schemes for the evolution of vertebrate immune systems (e.g., Stewart 1992), because it shows that the origins of diverse T-cell characteristics, such as antigen recognition and effector functions, may be separate, independent evolutionary events. Continued phylogenetic analysis of TCR and Ig relationships, especially as more suitable outgroup sequences are obtained from organisms such as cyclostomes or sharks, should continue to improve our understanding of immune system evolution." (p. 154)The reference to Stewart is: Stewart, J. 1992. Immunoglobulins did not arise in evolution to fight infection. Immunology Today 13:396-399. Stewart is also the author of the 1994 book The Primordial VRM System and the Evolution of Vertebrate Immunity.
Roth, D. B. (2000). "From lymphocytes to sharks: V(D)J recombinase moves to the germline." Genome Biology 1(2): 1014.1011-1014.1014. (PubMed | DOI | Journal | Google Scholar)
This article reviews the transposon model for the origin of rearranging receptors, and also explores the impact that the V(D)J recombinase may have had as an evolutionary force."Early on, it was suggested that the V(D)J recombination system might have arisen by the fortuitous integration of a transposable element into an ancestral antigen-receptor gene [14]. This hypothesis was strengthened by the discovery that the RAG genes are tightly linked [7], and by the finding that the RAG proteins can act as a transposase. Thus, a plausible model for the acquisition of the V(D)J recombination system during vertebrate evolution is the integration of a transposable element carrying the linked RAG genes into a primordial antigen-receptor gene in an ancestral jawed vertebrate, approximately 450 million years ago (reviewed in [1,11]). Presumably, this initial integration event created the first rearranging antigen-receptor gene; subsequent gene duplication events then created the multiple immunoglobulin and T-cell receptor loci." (p. 1014.2)
[Notable References]
1. Thompson CB: New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity 1995, 3:531-539.
2. Litman GW, Anderson MK, Rast JP: Evolution of antigen binding receptors. Annu Rev Immunol 1999, 17:109-147.
3. Marchalonis JJ, Schluter SF, Bernstein RM, Shen S, Edmundson AB: Phylogenetic emergence and molecular evolution of the immunoglobulin family. Adv Immunol 1998, 70:417-506.
4. Lee SS, Fitch D, Flajnik MF, Hsu E: Rearrangement of immunoglobulin genes in shark germ cells. J Exp Med 2000, 191:1637-1648.
[...]
7. Oettinger MA, Schatz DG, Gorka C, Baltimore D: RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 1990, 248:1517-1523.
[...]
11. Hansen JD, McBlane JF: Origin and evolution of the vertebrate immune system. Curr Topics Microbiol Immunol 2000, 248:111-135.
12. Agrawal A, Eastman QM, Schatz DG: Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 1998, 394:744-751.
13. Hiom K, Melek M, Gellert M: DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 1998, 94:463-470.
14. Sakano H, Huppi K, Heinrich G, Tonegawa S: Sequences at the somatic recombination sites of immunoglobulin light chain genes. Nature 1979, 280:288-294.
Vaandrager, J.-W., Schuuring, E., Philippo, K. and Kluin, P. M. (2000). "V(D)J recombinase-mediated transposition of the BCL2 gene to the IGH locus in follicular lymphoma." Blood 96(5): 1947-1952. (PubMed | Journal | Google Scholar)
The results of this research article give indirect evidence that transposition reactions mediated by RAGs can occur not just in vitro, but within mammalian cells, further supporting the transposon hypothesis.
Du Pasquier, L. (2001). "The immune system of invertebrates and vertebrates." Comparative Biochemistry and Physiology Part B Biochemistry & Molecular Biology 129(1): 1-15. (PubMed | DOI | Journal | Google Scholar)
Review of current comparative immunology. Excellent figures show the variability and repeated themes (due to duplication and combination of genes) in various immune systems. Fig. 3 shows the repeated use of proteins in the immunoglobulin superfamily in invertebrates.
Figure 2 mentions the transposon hypothesis:"The introduction of what nowadays behaves as a site sensitive to the RAG enzyme is supposed to have occurred via the introduction of a transposon (Rougeon, 1986; Thompson, 1995)." (p. 5)The text does also:"It is as if a 'horizontal' acquisition of a transposon had occurred at some point during the history of the vertebrates (Thompson, 1995). The receptor gene structure itself, in the region where the rearrangement takes place, suggests the introduction of a transposon. The recombination signal sequences flank the gene segments (V and J in the light chain for instance) in the same way as the LTR does in a transposon (Hansen and McBlane, 2000). In principle, RAG activity is confined to lymphocytes but the existence of germ-line rearrangements in light chain genes in the shark suggests that RAG (Lee et al., 2000) is still an active force changing the genome in some vertebrates."
[References]
Hansen, J.D., McBlane, J.F., 2000. Recombination-activating genes, transposition, and the lymphoid-specific combinatorial immune system: a common evolutionary connection (In Process Citation). Curr. Top. Microbiol. Immunol. 248, 111-135.
Lee, S.S., Fitch, D., Flajnik, M.F., Hsu, E., 2000. Rearrangement of immunoglobulin genes in shark germ cells (see comments). J. Exp. Med. 191, 1637-1648.
Rougeon, F., 1986. La diversité des anticorps. La Recherche 17, 680-689.
Thompson, C.B., 1995. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity 3, 531-539.
Nonaka, M. and Miyazawa, S. (2001). "Evolution of the Initiating Enzymes of the Complement System." Genome Biology 3(1): 1001.1001-1001.1005. (PubMed | DOI | Journal | Google Scholar)
Technical review of the origin of the innate (non-adaptive) immune system.
"Figure 2 A proposal for the evolution of the MASP-113, MASP-2, C1r and C1s genes." (p. 1001.3)"The primitive complement system
As discussed here, the complement system seems to have a more ancient origin in evolution than adaptive immunity; the latter seems to have been established only from jawed vertebrates onwards. The central component of the complement system, the C3 protein on which the three activation pathways discussed here converge, has been identified in jawless vertebrates, the lamprey [13] and hagfish [14], as well as in deuterostome invertebrates, amphioxus (a cephalochordate) [15], ascidian (urochordate) [16] and sea urchin (echinoderm) [17]. Although C3- or complement-like molecules of insects show functional similarity with deuterostome C3, there is no shared derived structural character between them, indicating that the deuterostome C3 and insect C3-like molecules derived independently from their common ancestor, [alpha]2-macroglobulin. Thus, the authentic complement system seems to have been established in the deuterostome lineage; the classical pathway of complement activation was then acquired in the jawed vertebrate lineage, at the time adaptive immunity arose." (p. 1001.4)
Cannon, J. P., Haire, R. N. and Litman, G. W. (2002). "Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate." Nature Immunology 3(12): 1200-1207. (PubMed | DOI | Journal | Google Scholar)
A key research article finally presenting evidence of a non-rearranging receptor closely related to the vertebrate rearranging receptors, in the lancelet (a cephalochordate which lacks adaptive immunity). This is important because it provides one of the homologs that scientists had been seeking for years. It also shows that even without rearranging receptor capability, selection favored diverse receptors:"Thus, multigenic families encoding diversified V regions exist in a species lacking an adaptive immune response." (p. 1200)
Kaufman, J. (2002). "The origins of the adaptive immune system: whatever next?" Nature Immunology 3(12): 1124-1125. (PubMed | DOI | Journal | Google Scholar)
In this News & Views article in Nature Immunology, Kaufman expresses wonderment at the results of Cannon et al. (2002)."Indeed, the initial event that enabled the emergence of the adaptive immune system is thought to have been the insertion of a transposon into a previously undisrupted V-like exon of an Ig domain-containing gene2,3,5. But in which Ig gene did the insertion occur?" (p. 1124)
"The obvious approach to answer these questions is to search for a V-like Ig-containing gene in a jawless vertebrate or a nonvertebrate chordate." (p. 1124)
"The discovery of a V-like Ig multigene family in the protochordate amphioxus provides new insights into the evolution of the adaptive immune response." (p. 1124, summary)
[References]
1. Cannon, J. P., Haire, R. N. & Litman, G. L. Nature Immunol. 3, 1200-1207 (2002).
2. Litman, G. W., Anderson, M. K., & Rast, J. P., Annu. Rev. Immunol. 17, 109-147 (1999).
3. Flajnik, M. F. & Kasahara, M. Immunity 15, 351-362 (2001).
[...]
5. Tonegawa, S. Nature 302, 575-581 (1983).
Krem, M. M. and Di Cera, E. (2002). "Evolution of enzyme cascades from embryonic development to blood coagulation." Trends in Biochemical Sciences 27(2): 67-74. (PubMed | DOI | Journal | Google Scholar)
Article reviews the evolutionary and physiological relationships between the blood-clotting cascade and the complement cascade of the innate immune system."Here, it is proposed that a single ancestral developmental/immunity cascade gave rise to the protostome and deuterostome developmental, clotting, and complement cascades. Extensive similarities suggest that these cascades were built by adding enzymes from the bottom of the cascade up and from similar macromolecular building blocks." (p. 67)Figure 2 (p. 71) shows proposed relationships."Fig. 2. Evolutionary changes producing developmental, immune and clotting cascades."
"The emergence of the advanced deuterostome complement system, featuring both the classical and alternative pathways, occurred by duplication of primitive complement proteins and recruitment of new proteases from the pool of ancestral building blocks." (p. 73)
"The modern mammalian clotting cascade was completed by the addition of serine proteases such as factors XI and XII, and cofactors such as factors V and VIII, which conferred additional levels of positive and negative feedback regulation." (p. 73)
Laird, D. J. (2002). "Immune System." Encyclopedia of Evolution. Edited by M. Pagel. Oxford, Oxford University Press. 2: 558-564. (Library | Publisher | Amazon | Google Print)
Basic introduction to the immune system, but in a thoroughly evolutionary context. Figure 4 (p. 563) depicts the progressive aquisition of features as one moves closer to mammals in the vertebrate phylogeny. It is noteworthy that the transposon hypothesis is sufficiently well-accepted, and well-known, to make it into a general encyclopedia on evolution. Furthermore, this is another example of the secondary literature that is readily available to anyone seriously looking into evolutionary immunology."Structures of the Vertebrate Immune System. Despite the quantity of immune receptors, signalling molecules, and serum factors that carry out antigen recognition and the accompanying inflammatory response, these structures share much in common. Protein motifs such as the immunoglobulin fold, which appears in Ig, TCR, natural killer (NK) cell receptors, MHC receptors, complement proteins, and adhesion molecules, imply extraordinary recycling and even a common origin during evolution. The hypothesis that a primordial immunoglobulinlike receptor gave rise to the rearranging receptors of lymphocytes has inspired the study of Ig, TCR, and other immune receptors of primitive vertebrates. This section focuses on the evolution of the main components of the adaptive immune system, unique to jawed vertebrates." (p. 559)
"The sudden appearance of RAG in Chondrychthyes, coupled with their peculiar genetic features (they lack introns and are adjacent in most species) suggests their origin as a retrotransposon, or a foreign DNA element introduced by a virus some 500 million years ago." (p. 561)
"The origin of adaptive immunity can be approached from many angles. The genomic approach was first employed by Masanori Kasahara, who hypothesized that genome duplications in vertebrate ancestors paved the way for the molecular experimentation that gave rise to this system. Comparison of MHC loci across vertebrate species provides clues about how and when this chromosomal region of immune and nonimmune genes was assembled. The study of RAG structure and mechanism across vertebrates fuels the theory that receptor genes of the adaptive immune system first arose by transposon-induced shuttling of duplicated segments of DNA. Phylogenetic data estimate the appearance of RAG in a vertebrate ancestor about 500 million years ago, approximately coincidental with the last hypothesized genome duplication and the origins of TCR, Ig, and MHC receptors. Another strategy is the reconstruction of the hypothetical ancestral receptor through discovery of surviving remnants, such as nonrearranging immunoglobulin domain receptors in vertebrates and invertebrates alike. Candidates include the paired Ig-like receptors, whose role as inhibitory and activating receptors for MHC I and MHC I-like molecules in mammals suggests their involvement in ancient self-nonself recognition. A number of other Ig-type nonrearranging receptors, such as the NAR in sharks and the NITR in pufferfish, contribute to our understanding of receptor evolution." (p. 564)
Marchalonis, J. J., Jensen, I. and Schluter, S. F. (2002). "Structural, antigenic and evolutionary analyses of immunoglobulins and T cell receptors." Journal of Molecular Recognition 15(5): 260-271. (PubMed | DOI | Journal | Google Scholar)
Long review of the work of the Marchalonis lab."This laboratory has focused upon the structure, function and evolution of antibodies and T cell receptors for more than 25 years." (p. 260)Evolution played a key role in the lab's study of antigen receptors. The article is interesting because it highlights the interrelationship between structural and evolutionary studies -- homology and phylogeny allow structural knowledge gained in one lineage (e.g., sharks) to be applied to another that is more medically relevant (such as humans). The authors also note that sharks were chosen as the study organisms of choice because any features conserved across the evolutionary distance between sharks and humans is likely to be functionally important:"We have studied antibodies, antibody production and cellular immunity in representatives of all the vertebrate classes, but we have chosen to concentrate our studies of antibody and T cell receptor structure on a comparison between the genes and molecules of sharks and humans because these two species represent the most evolutionarily divergent organisms to express the complete combinatorial immune system consisting of immunoglobulins, major histocompatability complex antigens, T cell receptors, and recombination activator genes (DuPasquier and Flajnik, 1999; Litman et al., 1999; Marchalonis and Schluter, 1998; Marchalonis et al., 1998a)." (p. 261)
Marchalonis, J. J., Kaveri, S., Lacroix-Desmazes, S. and Kazatchkine, M. D. (2002). "Natural recognition repertoire and the evolutionary emergence of the combinatorial immune system." FASEB Journal 16(8): 842-848. (PubMed | Journal | Google Scholar)
Technical review of several aspects of the origin of adaptive immunity, including the capacity for receptor diversity and its selective value, and the importance of clonal selection during maturation of specific antibodies. The quick review of the transposon hypothesis is quoted below:"The complete [Combinatorial Immune Response, CIR] ensemble is present in all jawed vertebrates (8-10). The system emerged rapidly and accidentally in evolution (3, 14, 18) probably as the result of horizontal transfer of microbial genes for site-specific recombination (4-6, 18, 19). Ab initio, the CIR was neither a defense mechanism nor part of an internal regulatory system (20), but its unparalleled capacity to generate recognition molecules in real time gave it the potential to function effectively in both contexts. The CIR co-opted preexisting cellular and humoral mechanisms for activation, differentiation, and destruction of bound targets including the NF[kappa]B pathway, the IL-1/TOLL receptor complex, and interaction with more ancient complement components to initiate cytolysis, killing, and trigger inflammation (12-15)." (p. 843)
[References]
3. Marchalonis, J. J., and Schluter, S. F. (1998) A stochastic model for the rapid emergence of specific vertebrate immunity incorporating horizontal transfer of systems enabling duplication and combinatorial diversification. J. Theo. Biol. 193, 429-444
4. Bernstein, R. M., Schluter, S. F., Bernstein, H., and Marchalonis, J. J. (1996) Primordial emergence of the recombination activating gene 1 (RAG1): Sequence of the complete shark gene indicates homology to microbial integrates. Proc. Natl. Acad. Sci USA 93, 9545-9459
5. Hiom, K., Melek, M., and Gellert, M. (1998) DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94, 463-470
6. Agrawal, A., Eastman, Q. M., and Schatz, D. G. (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature (London) 394, 744-751
7. Burnet, F. M. (1959) The Clonal Selection, Vanderbilt University Press, Nashville, Tennessee
8. Marchalonis, J. J., Schluter, S. F., Bernstein, R. M., and Hohman, V. S. (1998) Antibodies of sharks: revolution and evolution. Immunol. Rev. 166, 103-122
9. Litman, G. W., Anderson, M. K., and Rast, J. P. (1999) Evolution of antigen binding receptors. Annu. Rev. Immunol. 17, 109-147
10. DuPasquier, L., and Flajnik, M. (1999) Origin and evolution of the vertebrate immune system. In Fundamental Immunology (Paul, W. E., ed) pp. 605-650, Lippincott-Raven, Philadelphia
12. Fearon, D., and Locksley, R. M. (1996) The instructive role of innate immunity in the acquired immune response. Science 272, 50-53
13. Meister, M., Hetru, C., and Hoffman, J. A. (2000) The antimicrobial host defense of Drosophila. Curr. Top. Microbiol. Immunol. 248, 17-36
14. Marchalonis, J. J., and Schluter, S. F. (1990) On the relevance of invertebrate recognition and defense mechanisms to the emergence of the immune response of vertebrates. Scand. J. Immunol. 32, 13-20
15. Medzhitov, R., and Janeway, C. A. (1997) Innate Immunity: The virtues of a nonclonal system of recognition. Cell 91, 295-298
17. Good, R. A., and Papermaster, B. W. (1964) Ontogeny and phylogeny of adaptive immunity. Adv. Immunol. 4, 1-115
18. Schluter, S. F., Bernstein, R. M., Bernstein, H., and Marchalonis, J. J. (1999) 'Big Bang' emergence of the combinatorial immune system. Dev. Comp. Immunol. 23, 107-111
19. Plasterk, R. (1998) V(D)J recombination. Ragtime jumping. Nature (London) 394, 744-751
20. Stewart, J. (1992) Immunoglobulins did not arise in evolution to fight infection. Immunol. Today 13, 396-399
Clatworthy, A. E., Valencia, M. A., Haber, J. E. and Oettinger, M. A. (2003). "V(D)J recombination and RAG-mediated transposition in yeast." Molecular Cell 12(2): 489-499. (PubMed | DOI | Journal | Google Scholar)
This paper showed for the first time that RAG1 and RAG2 can act as a bona fide transposase in eukaryotic (yeast) cells, mediating the integration in the genome of an excised DNA fragment. The results confirmed another long-sought prediction of the transposon hypothesis:"Here, we show that expression in Saccharomyces cerevisiae of murine RAG1 and RAG2, the lymphoid-specific components of the V(D)J recombinase, is sufficient to induce V(D)J cleavage and rejoining in this lower eukaryote." (p. 489)
Flajnik, M. F., Miller, K. and Du Pasquier, L. (2003). "Evolution of the Immune System." Fundamental Immunology. Edited by W. E. Paul. Philadelphia, Lippincott Williams & Wilkins: 519-570. (Library | Amazon | Google Print)
This 51-page review of evolutionary immunology references 213 citations. Technical, but very useful tables and graphics showing how the major features of the vertebrate immune system are gradually acquired in organisms getting closer in the phylogenetic tree to vertebrates."As predicted over 10 years ago by Zasloff (2), it is informative from both intellectual and applied viewpoints to understand how all living things defend themselves; few would argue against the premise that studies of Drosophila humoral immunity has fueled great interest in innate immunity, both for its own sake and in the way it regulates adaptive immunity (R3)." (p. 521)Regarding the transposon hypothesis for the origin of the Recombination-Activating Genes, RAG1 and RAG2, Flajnik et al. write,"It seems that a 'horizontal' acquisition of a RAG-laden transposon occurred at some point during the history of the vertebrates (R156, 202). The structure of the receptor gene itself, in the region where rearrangements occur, suggests the introduction of a transposon." (p. 562)The article also suggests future research directions:"Further molecular studies in hagfish and lamprey and in non-vertebrate deuterostomes will perhaps permit a better understanding of the events leading to the emergence of the adaptive immune system." (p. 566)Note that in May 2005, a "free-living" transposon relative of RAG1 was discovered in a tunicate, a non-vertebrate deuterostome (Kapitonov and Jurka, 2005).
Just while this review article was being written, seven new research papers came out which the authors saw fit to mention in an addendum, "Publications Worth Noting."[References cited]
R2. Jacob L, Zasloff M. Potential therapeutic applications of magainins and other antimicrobial agents of animal origin. Ciba Found Symp 1994; 186:197-216.
R3. Janeway CA Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol 2002; 20:197-216.]
R156. Hansen, J. D., McBlane, J. F. Recombination-activating genes, transposition, and the lymphoid-specific combinatorial immune system: a common evolutionary connection. Current Topics in Microbiology and Immunology 2000; 248:111-135.
R202. Agrawal A., Eastman Q. M., Schatz D. G. Transposition mediated by RAG1 and RAG2 and its implication for the evolution of the immune system. Nature. 1998; 394:744-751.
Market, E. and Papavasiliou, F. N. (2003). "V(D)J Recombination and the Evolution of the Adaptive Immune System." PLoS Biology 1(1): 24-27. (PubMed | DOI | Journal | Google Scholar)
A short review of recombination, and its evolutionary origin from a transposon. Included in the very first issue (volume 1, number 1) of the journal Public Library of Science: Biology. The entire contents of this journal are available free online.
The article notes that RAGs are indispensable for receptor gene rearrangement -- section heading: "RAGs: Indispensable for V(D)J Recombination" (p. 26)
However, despite the claims of antievolutionists, it is clear that RAGs evolved anyway:"An Evolutionary Model of V(D)J Recombination
From the discovery of the RAG genes on, investigators have suspected that V(D)J recombination may be the result of the landing of a transposable genetic element (a "jumping gene" or transposon) into the vertebrate genome. The clues were many. Firstly, the compact organization of the RAG locus resembles a transposable element (Schatz et al. 1989). Secondly, RAGs cut the DNA after binding RSSs throughout the BCR and TCR loci (Gellert 2002). RSSs resemble the ends of other transposable elements. Biochemically, the reaction shares characteristics with enzymes found in other transposable elements (Gellert 2002). Finally, these genes appear abruptly in evolution: they are present in the jawed vertebrates (like the shark), but not in more ancient organisms (Schluter et al. 1999).
A current model is that an ancient transposon containing the RAG genes flanked by RSS ends "jumped’" into an area of the vertebrate lineage containing a primordial antigen receptor gene, separating it into pieces. When the transposon lifted off, it left the RSSs behind. Multiple rounds of transposition and duplication eventually gave rise to our TCR and BCR loci." (p. 27)
[References]
Gellert M (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem 71: 101-132.
Schatz DG, Oettinger MA, Baltimore D (1989) The V(D)J recombination activating gene, RAG-1. Cell 59: 1035-1048.
Schluter SF, Bernstein RM, Bernstein H, Marchalonis JJ (1999) 'Big Bang' emergence of the combinatorial immune system. Dev Comp Immunol 23: 107-111.
Messier, T. L., O'Neill, J. P., Hou, S.-M., Nicklas, J. A. and Finette, B. A. (2003). "In vivo transposition mediated by V(D)J recombinase in human T lymphocytes." The EMBO Journal 22(6): 1381-1388. (PubMed | DOI | Journal | Google Scholar)
Messier and coworkers show that in human T lymphocytes, the excised DNA fragments from VDJ recombination can be reintegrated in genomic DNA, in this case causing disruption of the gene for a metabolic enzyme. Like Clathworthy et al (2003, in this bibliography), this paper provides strong evidence for a role of RAG1/2 as a functional transposase.
Cannon, J. P., Haire, R. N., Rast, J. P. and Litman, G. W. (2004). "The phylogenetic origins of the antigen-binding receptors and somatic diversification mechanisms." Immunological Reviews 200(1): 12-22. (PubMed | DOI | Journal | Google Scholar)
Detailed review of the origin of adaptive immunity. It takes the transposon hypothesis for granted and deals mostly with the "setting" for the transposon insertion -- the ancestral receptors and cells, etc."A mechanism that most likely gave rise to the adaptive immune system has been identified (1, 2) and is discussed elsewhere in the text. However, to understand the evolution of vertebrate adaptive immunity in detail, the characteristics of the immune systems of pre-adaptive ancestors must be reconstructed.
[...]
1. Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 1998;394: 744-751.
2. Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 1998;94:463-470." (pp. 12-13, 21)
Davis, M. M. (2004). "The evolutionary and structural 'logic' of antigen receptor diversity." Seminars in Immunology 16: 239-243. (PubMed | DOI | Journal | Google Scholar)
Article on the evolutionary advantages of immune system receptor diversity. Highly technical.
Eason, D. D., Cannon, J. P., Haire, R. N., Rast, J. P., Ostrov, D. A. and Litman, G. W. (2004). "Mechanisms of antigen receptor evolution." Seminars in Immunology 16: 215-226. (PubMed | DOI | Journal | Google Scholar)
Detailed review of the evolutionary origin of adaptive receptors. All of the figures are useful summaries of various points.
The basic conclusion is that while the exact ancestral (pre-rearranging) receptor has not been found, many close relatives sharing key properties have been discovered."Taken together, the results suggest that the adaptive immune system in vertebrates may have been derived in unique ways from different sets of 'primordial' receptors, of which one gave rise to the Ig/TCR, another generated the VLRs, and other receptors provided protective immunity in invertebrates and protochordates, further blurring distinctions between the traditional views of adaptive and innate immunity." (pp. 224-225)
Flajnik, M. F. and Du Pasquier, L. (2004). "Evolution of innate and adaptive immunity: can we draw a line?" Trends in Immunology 25(12): 640-644. (PubMed | DOI | Journal | Google Scholar)
New developments in the study of the evolution of adaptive immunity, especially noting that the lines between "innate" and "adaptive" immunity are becoming blurred as more organisms are studied. Representative quotes:"Particularly exciting are the discoveries of a new gene rearrangement mechanism in lampreys and a somatic diversification of mollusk immune genes." (p. 640)
"For comparative immunologists, these new sequencing data have turned us into kids in a candy shop and have spawned new insights into the integration of different immune systems." (p. 640)
"The birth of the adaptive immune system is believed to have occurred when an Ig superfamily (Igsf) gene of the variable (V) type was invaded by a transposable element containing RAG1 and RAG2 genes [5,6]. This innovative event, which greatly augmented receptor diversity (and heightened the risk of autoimmunity), must have been crucial for the adaptive immune system, even if it were not truly the initiating event [7] because all of the antigen receptor loci rearrange in a similar fashion (see later). There are several invertebrate Igsf family members having features of Ig or TCR V genes, which could be related to the ancestral gene invaded by the transposon [8-10]." (p. 640, emphases original)
"Already, other V genes with features similar to Ig or the TCR and potentially related to the ancestral Igsf gene invaded by the transposon have been isolated...." (p. 641)
"Every immunology student is taught that the jawed-vertebrate adaptive immune system was grafted onto the innate system, co-opting pre-existing effector mechanisms of defense [27]." (p. 643)
[References]
1 Medzhitov, R. et al. (1997) A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394-397
2 Hoffmann, J.A. and Reichhart, J.M. (2002) Drosophila innate immunity: an evolutionary perspective. Nat. Immunol. 3, 121-126
3 G. Koretsky (ed. in chief) (2004) Primitive Immune Systems. (J.A. Hoffman, ed.) Immunol. Rev. 198, 303
4 Flajnik, M.F. et al. (2003) Evolution of the immune system. In Fundamental Immunology (Paul, W.E. ed.), pp. 519-570, Lippincott, Williams & Wilkins, Philadelphia
5 Bernstein, R.M. et al. (1996) Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc. Natl. Acad. Sci. U. S. A. 93, 9454-9459
6 Agrawal, A. et al. (1998) Transposition mediated by RAG1 and RAG and its implications for the evolution of the immune system. Nature 394, 744-751
7 Erwin, D.H. and Krakauer, D.C. (2004) Evolution. Insights into innovation. Science 304, 1117-1119
8 Azumi, K. et al. (2003) Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: "waiting for Godot". Immunogenetics 55, 570-581
9 Du Pasquier, L. et al. (2004) Immunoglobulin superfamily receptors in protochordates: before RAG time. Immunol. Rev. 198, 233-248
10 Van den Berg, T.K. et al. (2004) On the origins of adaptive immunity: innate immune receptors join the tale. Trends Immunol. 25, 11-16
Galaktionov, V. G. (2004). "Evolutionary Development of the Immunoglobulin Family." Biology Bulletin 31(2): 101-111. (PubMed | DOI | Journal | Google Scholar)
Reviews the evolutionary origin of immunoglobulins, particularly how the immunoglobulin domain has been reused in many different receptors."It is believed that RAG was originally introduced by a retrovirus into the genome of ancestral vertebrates, specifically, into the region occupied by the single V gene." (p. 110)
Gould, S. J., Hildreth, J. E. and Booth, A. M. (2004). "The evolution of alloimmunity and the genesis of adaptive immunity." Quarterly Review of Biology 79(4): 359-382. (PubMed | DOI | Journal | Google Scholar)
A long review article in the prestigious general biology journal Quarterly Review of Biology. Stephen J. Gould (a different one than the famous paleontologist) argues that alloimmunity (recognition of self vs. nonself) evolved first, allowing the slightly later evolution of the hypervariable adaptive immune system (the variability creates the risk of accidentally attacking self-cells, so preexisting alloimmunity would ameliorate this)."Given that alloimmunity arose more than 550 million years ago, at the dawn of animal evolution, and that adaptive immunity arose approximately 100 million years later at the origin of the jawed vertebrates (Schatz 1999), the most parsimonious explanation for why the two systems share so many features is that adaptive immunity evolved from a preexisting alloimmune system. We propose that hypervariable alloimmunity was exapted for adaptive immune responses soon after its genesis, an evolutionary event that could be selected by virtually any pathogen, even HAAPs." (p. 373)
Holmes, E. C. (2004). "Adaptation and Immunity." PLoS Biology 2(9): 1269-1269. (PubMed | DOI | Journal | Google Scholar)
This short primer article in PLoS Biology discusses the well-known adaptive value of having diverse antigen receptor genes, and the evolutionary arms race between immune system cells attempting to detect pathogens, and the pathogens attempting to avoid detection."It is therefore little wonder that the host and pathogen genes that control infection and immunity frequently show high levels of genetic diversity and present some of the best examples of positive selection (adaptive evolution) reported to date (Yang and Bilawski 2000). In particular, rates of nonsynonymous substitution per site (resulting in an amino acid change; dN) often greatly exceed those of synonymous substitution per site (silent change; dS), as expected if most mutations are fixed becuase they increase fitness." (p. 1267)
[Reference]
Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15: 496-502.
Jones, J. M. (2004). "The taming of a transposon: V(D)J recombination and the immune system." Immunological Reviews 200(1): 233-248. (PubMed | DOI | Journal | Google Scholar)
Review exploring the diversity of transposon systems and their relationship to V(D)J recombination."The study of V(D)J recombination has been continually informed by research into other systems, and it is now apparent that V(D)J recombination shares a particularly close relationship with the family of transposons that includes Ac/Ds. These similarities support the hypothesis that V(D)J recombination evolved from a primordial transposon." (pp. 233-234)
Nonaka, M. and Yoshizaki, F. (2004). "Evolution of the Complement System." Molecular Immunology 40(12): 897-902. (PubMed | DOI | Journal | Google Scholar)
Technical review of the origin of the innate (non-adaptive) immune system."Here, we discuss an early evolution of the complement components, both the non-modular C3 family and the other modular components, revealed by recent molecular analysis of possible complement genes from [a] variety of animals and the recently published genome analysis of an ascidian, [a tunicate or sea squirt], Ciona intestinalis." (p. 897)
"Thus, the evolution of the vertebrate complement system could be accomplished by integration of several independent functional units into a single reaction system through making a functional linkage among them." (p. 901)
Pancer, Z., Amemiya, C. T., Ehrhardt, G. R. A., Ceitlin, J., Gartland, G. L. and Cooper, M. D. (2004). "Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey." Nature 430(6996): 174-180. (PubMed | DOI | Journal | Google Scholar)
Evidence of a different system of adaptive immunity in an early-diverging vertebrate, the sea lamprey."Different evolutionary strategies were thus used to generate highly diverse lymphocyte receptors through rearrangement of LRR modules in agnathans (jawless fish) and of immunoglobulin gene segments in gnathostomes (jawed vertebrates)." (p. 174)
Rothenberg, E. V. and Pant, R. (2004). "Origins of lymphocyte developmental programs: transcription factor evidence." Seminars in Immunology 16(4): 227-238. (PubMed | DOI | Journal | Google Scholar)
Origins of lymphocytes (vertebrate immune system cells). Highly technical.
Cooption reference:"Co-option of group C factors from other ancestral roles appears to have been harnessed for control of TCR and Ig gene rearrangement and expression." (p. 234)
"The evidence we have reviewed indicates that the full range of different transcription factor types needed to make lymphocytes were available in the genome at least as early as the emergence of urochordates (e.g., ascidians). In other words, the existence of transcription factors of these general families was not the rate-limiting feature for the evolution of lymphocyte development." (p. 234)
Schatz, D. G. (2004). "Antigen receptor genes and the evolution of a recombinase." Seminars in Immunology 16(4): 245-256. (PubMed | DOI | Journal | Google Scholar)
Reviews the transposon hypothesis for the origin of rearranging receptors. Fig. 3 (p. 250) gives the RAG transposon model. The evidence is also reviewed in detail on p. 250.
The authors make an interesting prediction:"Hence, one might expect to find in the genome of vertebrates relics of these other jumps, including degenerate versions of pairs of RSSs or of the RAG open reading frames." (p. 251)
Stavnezer, J. and Amemiya, C. T. (2004). "Evolution of isotype switching." Seminars in Immunology 16(4): 257-275. (PubMed | DOI | Journal | Google Scholar)
Long, technical review of the evolution of an aspect of the adaptive immune system (CSR, class switch recombination) apparently restricted to tetrapods (amphibians, reptiles, mammals, birds)."This chapter will review the evolution of the ability to express antibodies, or immunoglobulins (Igs) of different classes or isotypes." (p. 257)Figure on p. 258 shows diversity of immune system gene organization.
Zhou, L., Mitra, R., Atkinson, P. W., Hickman, A. B., Dyda, F. and Craig, N. L. (2004). "Transposition of hAT elements links transposable elements and V(D)J recombination." Nature 432: 995-1001. (PubMed | DOI | Journal | Google Scholar)
Discovery of a transposon with mechanistic and sequence similarities to the RAG genes."It had been suggested that the V(D)J recombination system may have evolved from an ancient transposable DNA element12, 50. Our findings here of such a close mechanistic relationship between hAT transposition and V(D)J recombination...and the related active sites of hAT tranposases and RAG1 provides strong support for the view that V(D)J recombination evolved from transposable elements." (p. 1000)
[References]
12. Gellert, M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71, 101-132 (2002).
18. Hiom, K., Melek, M. & Gellert, M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94, 463-470 (1998).
19. Agrawal, A., Eastman, O. M. & Schatz, D. G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744-751 (1998).
20. van Gent, D. C., Mizuuchi, K. & Gellert, M. Similarities between initiation of V(D)J recombination and retroviral integration. Science 271, 1592-1594 (1996).
50. Lewis, S. M. & Wu, G. E. The origins of V(D)J recombination. Cell 88, 159-162 (1997).
Klein, J. and Nikolaidis, N. (2005). "The descent of the antibody-based immune system by gradual evolution." Proceedings of the National Academy of Sciences 102(1): 169-174. (PubMed | DOI | Journal | Google Scholar)
Argues against the "big bang" model of immune system evolution which asserts that the main features of the immune system came together "suddenly", i.e., in less than 50 million years. Instead, it argues that the processes was more graduated and step-by-step. Given that the research community seems to agree that the introduction of RAG clearly was a key step and would be "sudden" compared to other forms of genetic change, but also agrees that many other more common evolutionary processes and precursors were also involved in the origin of adaptive immunity, it can be argued that debating the "big bang" label is not particularly illuminating. Regardless, in doing so Klein and Nikolaidis re-emphasize some useful points:"We argue that the AIS [Antibody-based Immune System] has been assembled from elements that have primarily evolved to serve other functions and incorporated existing molecular cascades, resulting in the appearance of new organs and new types of cells." (p. 169)Figure 2 shows how many organisms lack many of the parts found in the most sophisticated systems.
"In reality, however, the AIS is an example of an organ system that has evolved gradually through a series of small steps over an extended period." (p. 169)
Cannon, J. P., Haire, R. N., Pancer, Z., Mueller, M. G., Skapura, D., Cooper, M. D. and Litman, G. W. (2005). "Variable Domains and a VpreB-like molecule are present in a jawless vertebrate." Immunogenetics 56(12): 924-929. (PubMed | DOI | Journal | Google Scholar)
This paper describes a family of non-rearranging receptors in sea lampreys that are closely related to modern immunoglobulins and, together with papers by Cannon et al (2002, in this bibliography) and Suzuki et al (2005, Journal of Immunology, 174:2885-91) constitutes strong evidence of the existence of multiple, diverse families of immunoglobulin-like proteins predating the rearranging antigen receptors found in jawed vertebrates."This is the first indication of a molecule related to the B cell receptor (BCR) complex in a species that diverged prior to the jawed vertebrates in which RAG-mediated adaptive immunity is first encountered." (p. 924)
Kapitonov, V. V. and Jurka, J. (2005). "RAG1 Core and V(D)J Recombination Signal Sequences Were Derived from Transib Transposons." Public Library of Science Biology 3(6): e181:0001-0014. (PubMed | DOI | Journal | Google Scholar)
Reports the discovery of homologs of RAG1 in several non-chordate phyla -- this is the direct evidence of a free-living transposon suggested by Lewis in 1999. The summary figure was used by Ken Miller during his trial testimony."Even prior to the discovery of RAG1 and RAG2, it had been suggested that the first two RSSs were originally terminal inverted repeats (TIRs) of an ancient transposon whose accidental insertion into a gene ancestral to BCR and TCR, followed by gene duplications, triggered the emergence of the V(D)J machinery [4]. Later, this model was expanded by the suggestion that both RAG1 and RAG2 might have evolved from a transposase (TPase) that catalyzed transpositions of ancient transposons flanked by TIRs that were precursors of RSSs [9]. This model has received additional support through observations of similar biochemical reactions in transposition and V(D)J recombination [10,11]. Finally, it was demonstrated that RAG1/2 catalyzed transpositions of a DNA segment flanked by RSS12 and RSS23 in vitro [12,13] and in vivo in yeast [14]. In vertebrates, in vivo RAG-mediated transpositions are strongly suppressed, probably to minimize potential harm to genome function. So far, only one putative instance of such a transposition has been reported [15]. However, given the lack of significant structural similarities between RAGs and known TPases, the "RAG transposon" model [9,12,13,16] remained unproven. Here we demonstrate that the RAG1 core and RSSs were derived from a TPase and TIRs encoded by ancient DNA transposons from the Transib superfamily [17]." (pp. e181-e182)
[References]
4. Sakano H, Huppi K, Heinrich G, Tonegawa S (1979) Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280: 288-294.
[...]
9. Thompson CB (1995) New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity 3: 531-539.
10. van Gent DC, Mizuuchi K, Gellert M (1996) Similarities between initiation of V(D)J recombination and retroviral integration. Science 271: 1592-1594.
11. Melek M, Gellert M, van Gent DC (1998) Rejoining of DNA by the RAG1 and RAG2 proteins. Science 280: 301-303.
12. Agrawal A, Eastman QM, Schatz DG (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394: 744-751.
13. Hiom K, Melek M, Gellert M (1998) DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell 94: 463-470.
14. Clatworthy AE, Valencia MA, Haber JE, Oettinger MA (2003) V(D)J recombination and RAG-mediated transposition in yeast. Mol Cell 12: 489-499.
15. Messier TL, O'Neill JP, Hou SM, Nicklas JA, Finette BA (2003) In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J 22: 1381-1388.
16. Lewis SM, Wu GE (2000) The old and the restless. J Exp Med 191: 1631-1636.
17. Kapitonov VV, Jurka J (2003) Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A 100: 6569-6574.
Du Pasquier, L. (2005). "Meeting the demand for innate and adaptive immunities during evolution." Scandinavian Journal of Immunology 62(s1): 39-48. (PubMed | DOI | Journal | Google Scholar)
Detailed review of the total immune responses across metazoa, focusing on the demands imposed by natural selection, and the evolution of cooperation between adaptive and nonadaptive systems."Immunoglobulins and T-cell receptors
The RAG1 and 2 complexes looked so far like the only new component responsible for the adaptive system of vertebrates because no related genes were found in invertebrates. However, the discovery of a RAG2-like product with homologs among metazoa and plants (called 'peas' and showing the same composition in 'kelch' domains as RAG2) and associated with the meiotic chromatin, may force us to revise the hypothesis of a RAG transfer by transposition in the immediate ancestors of gnathostomes [32]. What is in question is perhaps not the transposing-like feature of this enzymatic system but the way it was involved in immunity and its association with RAG1. Could have the lymphocyte RAG2 arisen by duplication of a 'peas'-like gene at the time of genome duplications? Maybe the introduction of RAG1 of which [a] homolog can now be found in Echinoderms [33] was the key step for the adaptive system." (p. 44)
"In the Introduction, the simultaneity and the diversity of the pressures that led to the development of immunity at the beginning of the existence of metazoa were stressed. As a result, independent pathways of immunity functioning on different principles of recognition developed, probably in parallel, each being pushed for diversification during adaptation to the changing environment. Some organisms trusted conservatively the genes devoted to their immune system; other added and allowed some randomness in their expression. In the comparative studies one has observed convergences, mimicry, duplications, dead ends, complementation or even perhaps competitions, with a trend towards individualization of responses to the point where the distinctions between adaptive and innate immunity are uncertain. The diversification of the immune molecules has made the necessary control of their expression a difficult task, demanding the involvement of many other gene families that were recruited alongside in the build up of the integrated immune systems. The individual defense mechanisms do not work in isolation in any organism but instead are part of a coherent whole. In fact, there are many unknown genes expressed during immune responses of the simplest organisms outside vertebrates [54]. In addition to the now more fashionable protochordates and jaw-less vertebrates, it would be important to include the study of arthropods less derived than Drosophila such as the horseshoe crab. One should also include the Cnidaria whose genes’ sequences are surprisingly close to those of deuterostomes and the genome of which could contain a reservoir of interesting information for the immunologist preoccupied with evolutionary issues [55]. In those organisms, examination of the genome and individual studies of the regulations and interactions of the different components of their immune systems may provide information as to how to handle the dysfunctions of what I foolishly dared to call in the introduction our ideal Homo sapiens immune system." (p. 47)
[References]
32 Ohinata Y, Sutou S, Mitsui Y. A novel testis-specific RAG2-like protein, Peas: its expression in pachytene spermatocyte cytoplasm and meiotic chromatin. FEBS Lett 2003;537:1-5.
33 Cannon JP, Haire RN, Rast JP, Litman GW. The phylogenetic origins of the antigen-binding receptors and somatic diversification mechanisms. Immunol Rev 2004;200:12-22.
54 De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genomewide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA 2001;98:12590-5.
55 Galliot B, Schmid V. Cnidarians as a model system for understanding evolution and regeneration. Int J Dev Biol 2002;46 (1 Spec No): 39-48.
Janssen, B. J. C., Huizinga1, E. G., Raaijmakers, H. C. A., Roos, A., Daha, M. R., Nilsson-Ekdahl, K., Nilsson, B. and Gros, P. (2005). "Structures of complement component C3 provide insights into the function and evolution of immunity." Nature 437(7058): 505-511. (PubMed | DOI | Journal | Google Scholar)
The authors report the crystal structure of the key innate immunity protein, C3, and deduce certain features of the evolutionary origin of the protein, such as internal domain duplications. This research came out the week before trial and was cited in Kenneth Miller's direct testimony.
Cohn, M. (2006). "What are the commonalities governing the behavior of humoral immune recognitive repertoires?" Developmental and Comparative Immunology 30(1-2): 19-42. (PubMed | DOI | Journal | Google Scholar)
Long review of selection-related issues in the evolution of immunity."The evolution of the immune system has been reviewed over the years from several points of view [1-10]. I will try to take a different tack by stressing selectable function rather than structure, genealogy, sequence, homology, gene dynamics, etc., the basic tools of comparative immunologists. This is not to downplay the importance of these analyses, which are, in fact, fundamental and indispensable, but rather to see whether another approach will confirm our present views or, hopefully, give us new insights." (p. 19)
[References]
[1] Du Pasquier L, Flajnik M. Origin and evolution of the vertebrate immune system. In: Paul WE, editor. Fundamental immunology. Philadelphia, PA: Lippincott-Raven Publishers; 1999. p. 605-50.
[2] Diaz M, Flajnik MF. Evolution of somatic hypermutation and gene conversion in adaptive immunity. Immunol Rev 1998; 162:13-24.
[3] Butler JE. Immunoglobulin gene organization and the mechanism of repertoire development. Scand J Immunol 1997;45:455-62.
[4] Du Pasquier L, Wilson M, Greenberg AS, Flajnik MF. Somatic mutation in ectothermic vertebrates: Musings on selection and origins. Curr Topics Microbiol Immunol 1998; 229:199-216.
[5] Flajnik M, Rumfelt LL. Early and natural antibodies in nonmammalian vertebrates. Curr Topics Microbiol Immunol 2000;252:233-40.
[6] Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol 1999;17:109-47.
[7] Immune systems of ectothermic vertebrates. In: Parham P, editor. Immunological reviews, 1998. p. 166-384.
[8] Flajnik M. Comparative analysis of immunoglobulin genes: Surprises and portents. Nat Rev Immunol 2002;2:688-98.
[9] Marchalonis JJ, Jensen I, Schuluter SF. Structural, antigenic and evolutionary analyses of immunoglobulins and T cell receptors. J Mol Recogn 2002;15:260-71.
[10] Neuberger MS. Antibodies: a paradigm for the evolution of molecular recognition. Biochem Soc Trans 2002;30:341-50.
Marchalonis, J. J., Adelman, M. K., Schluter, S. F. and Ramsland, P. A. (2006). "The antibody repertoire in evolution: Chance, selection, and continuity." Developmental and Comparative Immunology 30(1-2): 223-247. (PubMed | DOI | Journal | Google Scholar)
Massive review with 177 references. Deals with mutation and selection explicitly."At the onset, we emphasize that evolution is a stochastic or accidental process building upon the spontaneous generation of mutations followed by natural selection based upon survival to reproduce under external/environmental conditions." (p. 224)
"It has been proposed that the rapid evolutionary emergence of the combinatorial response was catalyzed by the horizontal transfer of RAG genes to an ancestral vertebrate [22-26]. The prokaryotic origin of these genes is buttressed by the lack of introns and by homology to microbial site-specific recombinases [23,27-29]." (p. 224)
[References]
[22] Bartl S, Baltimore D, Weissman IL. Molecular evolution of the vertebrate immune system. Proc Natl Acad Sci USA 1994;91:10769-70.
[23] Bernstein RM, Schluter SF, Bernstein H, Marchalonis JJ. Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc Natl Acad Sci USA 1996;93:9454-9.
[24] Schluter SF, Bernstein RM, Bernstein H, Marchalonis JJ. ‘Big Bang‘ emergence of the combinatorial immune system. Dev Comp Immunol 1999;23:107-11.
[25] Laird D, De Tomaso A, Cooper MD, Weissman I. 50 million years of chordate evolution: seeking the origins of adaptive immunity. Proc Natl Acad Sci USA 2000;97:6924-6.
[26] Plasterk R. V(D)J recombination. Ragtime jumping. Nature 1998;394:718-9.
[27] Banerjee-Basu S, Baxevanis AD. The DNA-binding region of RAG1 is not a homoeodomain. Genome Biol 2002;3:1004.
[28] Schatz DG. Transposition mediated by RAG1 and RAG2 and the evolution of the adaptive immune system. Immunol Res 1999;19:169-82.
[29] Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 1998;394:744-51.
